+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1578 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacillus subtilis RNA polymerase NusA complex | |||||||||
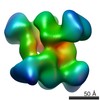
 Map data Map data | RNAP_NusA complex from Bacillus subtilis | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNAP NusA Transcription elongation Firmicutes | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Yang X / Molimau S / Doherty GP / Marles-Wright J / Rothnagel R / Hankamer B / Lewis RJ / Lewis PJ | |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2009 Journal: EMBO Rep / Year: 2009Title: The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. Authors: Xiao Yang / Seeseei Molimau / Geoff P Doherty / Elecia B Johnston / Jon Marles-Wright / Rosalba Rothnagel / Ben Hankamer / Richard J Lewis / Peter J Lewis /  Abstract: There are three stages of transcribing DNA into RNA. These stages are initiation, elongation and termination, and they are well-understood biochemically. However, despite the plethora of structural ...There are three stages of transcribing DNA into RNA. These stages are initiation, elongation and termination, and they are well-understood biochemically. However, despite the plethora of structural information made available on RNA polymerase in the last decade, little is available for RNA polymerase in complex with transcription elongation factors. To understand the mechanisms of transcriptional regulation, we describe the first structure, to our knowledge, for a bacterial RNA polymerase in complex with an essential transcription elongation factor. The resulting structure formed between the RNA polymerase and NusA from Bacillus subtilis provides important insights into the transition from an initiation complex to an elongation complex, and how NusA is able to modulate transcription elongation and termination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1578.map.gz emd_1578.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1578-v30.xml emd-1578-v30.xml emd-1578.xml emd-1578.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  1578.gif 1578.gif | 48.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1578 http://ftp.pdbj.org/pub/emdb/structures/EMD-1578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1578 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1578 | HTTPS FTP |
-Validation report
| Summary document |  emd_1578_validation.pdf.gz emd_1578_validation.pdf.gz | 230.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1578_full_validation.pdf.gz emd_1578_full_validation.pdf.gz | 229.3 KB | Display | |
| Data in XML |  emd_1578_validation.xml.gz emd_1578_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1578 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1578 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1578 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1578.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1578.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNAP_NusA complex from Bacillus subtilis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RNAP_NusA complex
| Entire | Name: RNAP_NusA complex |
|---|---|
| Components |
|
-Supramolecule #1000: RNAP_NusA complex
| Supramolecule | Name: RNAP_NusA complex / type: sample / ID: 1000 / Details: Monodisperse / Oligomeric state: Dimeric / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 440 KDa / Theoretical: 440 KDa / Method: Gel filtration |
-Macromolecule #1: RNA polymerase
| Macromolecule | Name: RNA polymerase / type: protein_or_peptide / ID: 1 / Name.synonym: RNAP / Details: Negative staining / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 400 KDa / Theoretical: 400 KDa |
-Macromolecule #2: NusA
| Macromolecule | Name: NusA / type: protein_or_peptide / ID: 2 / Name.synonym: NusA / Details: Negative staining / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 40 KDa / Theoretical: 40 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Details: 20 mM Tris-HCl pH 7.8, 150 mM NaCl, 10 mM MgCl2, 5% (v/v) glycerol and 1 mM DTT |
|---|---|
| Staining | Type: NEGATIVE / Details: 1% Uranyl acetate |
| Grid | Details: Home made carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Digitization - Sampling interval: 3.9 µm / Number real images: 524 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 15027 |
|---|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)