+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PAPP-A dimer in complex with a dimer of the inhibitor STC2 | |||||||||
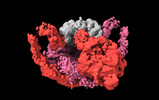
 Map data Map data | Primary, sharpened map of PAPP-A dimer with its endogenous inhibitor STC2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Metzincin metalloprotease Inhibitor complex / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of hormone biosynthetic process / regulation of store-operated calcium entry / response to vitamin D / negative regulation of multicellular organism growth / decidualization / endoplasmic reticulum unfolded protein response / embryo implantation / Post-translational protein phosphorylation / hormone activity / response to peptide hormone ...regulation of hormone biosynthetic process / regulation of store-operated calcium entry / response to vitamin D / negative regulation of multicellular organism growth / decidualization / endoplasmic reticulum unfolded protein response / embryo implantation / Post-translational protein phosphorylation / hormone activity / response to peptide hormone / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / intracellular calcium ion homeostasis / response to oxidative stress / cellular response to hypoxia / endoplasmic reticulum lumen / negative regulation of gene expression / heme binding / perinuclear region of cytoplasm / enzyme binding / endoplasmic reticulum / Golgi apparatus / protein homodimerization activity / extracellular space Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
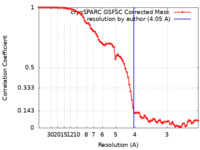
| Method | single particle reconstruction / cryo EM / Resolution: 4.05 Å | |||||||||
 Authors Authors | Kobbero SD / Oxvig C / Gajhede M / Boesen T | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism. Authors: Sara Dam Kobberø / Michael Gajhede / Osman Asghar Mirza / Søren Kløverpris / Troels Rønn Kjær / Jakob Hauge Mikkelsen / Thomas Boesen / Claus Oxvig /  Abstract: The metzincin metalloproteinase PAPP-A plays a key role in the regulation of insulin-like growth factor (IGF) signaling by specific cleavage of inhibitory IGF binding proteins (IGFBPs). Using single- ...The metzincin metalloproteinase PAPP-A plays a key role in the regulation of insulin-like growth factor (IGF) signaling by specific cleavage of inhibitory IGF binding proteins (IGFBPs). Using single-particle cryo-electron microscopy (cryo-EM), we here report the structure of PAPP-A in complex with its endogenous inhibitor, stanniocalcin-2 (STC2), neither of which have been reported before. The highest resolution (3.1 Å) was obtained for the STC2 subunit and the N-terminal approximately 1000 residues of the PAPP-A subunit. The 500 kDa 2:2 PAPP-A·STC2 complex is a flexible multidomain ensemble with numerous interdomain contacts. In particular, a specific disulfide bond between the subunits of STC2 and PAPP-A prevents dissociation, and interactions between STC2 and a module located in the very C-terminal end of the PAPP-A subunit prevent binding of its main substrate, IGFBP-4. While devoid of activity towards IGFBP-4, the active site cleft of the catalytic domain is accessible in the inhibited PAPP-A·STC2 complex, as shown by its ability to hydrolyze a synthetic peptide derived from IGFBP-4. Relevant to multiple human pathologies, this unusual mechanism of proteolytic inhibition may support the development of specific pharmaceutical agents, by which IGF signaling can be indirectly modulated. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15217.map.gz emd_15217.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15217-v30.xml emd-15217-v30.xml emd-15217.xml emd-15217.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15217_fsc.xml emd_15217_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15217.png emd_15217.png | 72 KB | ||
| Filedesc metadata |  emd-15217.cif.gz emd-15217.cif.gz | 6.2 KB | ||
| Others |  emd_15217_additional_1.map.gz emd_15217_additional_1.map.gz emd_15217_half_map_1.map.gz emd_15217_half_map_1.map.gz emd_15217_half_map_2.map.gz emd_15217_half_map_2.map.gz | 31.6 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15217 http://ftp.pdbj.org/pub/emdb/structures/EMD-15217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15217 | HTTPS FTP |
-Related structure data
| Related structure data |  8a7dC  8a7eC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15217.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15217.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary, sharpened map of PAPP-A dimer with its endogenous inhibitor STC2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.18594 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Raw map of PAPP-A dimer with its endogenous inhibitor STC2
| File | emd_15217_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map of PAPP-A dimer with its endogenous inhibitor STC2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
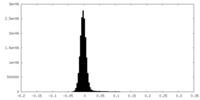
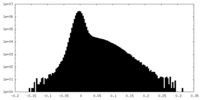
| Density Histograms |
-Half map: Half map A from the raw map
| File | emd_15217_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from the raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
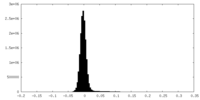
| Density Histograms |
-Half map: Half map B from the raw map
| File | emd_15217_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from the raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
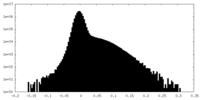
| Density Histograms |
- Sample components
Sample components
-Entire : PAPP-A dimer in complex with its endogenous inhibitor STC2 dimer
| Entire | Name: PAPP-A dimer in complex with its endogenous inhibitor STC2 dimer |
|---|---|
| Components |
|
-Supramolecule #1: PAPP-A dimer in complex with its endogenous inhibitor STC2 dimer
| Supramolecule | Name: PAPP-A dimer in complex with its endogenous inhibitor STC2 dimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Inhibited proteolytic complex generated by harvest of serum media, purifying on a nickel column followed by negative affinity purification and size-exclusion chromatography. |
|---|---|
| Molecular weight | Theoretical: 400 KDa |
-Supramolecule #2: PAPP-A
| Supramolecule | Name: PAPP-A / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: Homodimer Pregnancy-associated plasma protein A |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: ubiquitous / Location in cell: extracellular Homo sapiens (human) / Tissue: ubiquitous / Location in cell: extracellular |
-Supramolecule #3: Stanniocalcin-2
| Supramolecule | Name: Stanniocalcin-2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 / Details: Homodimer |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: ubiquitous / Location in cell: extracellular Homo sapiens (human) / Tissue: ubiquitous / Location in cell: extracellular |
-Macromolecule #1: PAPP-A
| Macromolecule | Name: PAPP-A / type: protein_or_peptide / ID: 1 Details: A heterotetrameric complex between the protease PAPP-A dimer and its endogenous inhibitor STC2 Enantiomer: LEVO / EC number: pappalysin-1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EARGATEEPS PPSRALYFSG RGEQLRLRAD LELPRDAFT LQVWLRAEGG QRSPAVITGL YDKCSYISRD RGWVVGIHTI SDQDNKDPRY FFSLKTDRAR QVTTINAHR SYLPGQWVYL AATYDGQFMK LYVNGAQVAT SGEQVGGIFS PLTQKCKVLM L GGSALNHN ...String: EARGATEEPS PPSRALYFSG RGEQLRLRAD LELPRDAFT LQVWLRAEGG QRSPAVITGL YDKCSYISRD RGWVVGIHTI SDQDNKDPRY FFSLKTDRAR QVTTINAHR SYLPGQWVYL AATYDGQFMK LYVNGAQVAT SGEQVGGIFS PLTQKCKVLM L GGSALNHN YRGYIEHFSL WKVARTQREI LSDMETHGAH TALPQLLLQE NWDNVKHAWS PM KDGSSPK VEFSNAHGFL LDTSLEPPLC GQTLCDNTEV IASYNQLSSF RQPKVVRYRV VNL YEDDHK NPTVTREQVD FQHHQLAEAF KQYNISWELD VLEVSNSSLR RRLILANCDI SKIG DENCD PECNHTLTGH DGGDCRHLRH PAFVKKQHNG VCDMDCNYER FNFDGGECCD PEITN VTQT CFDPDSPHRA YLDVNELKNI LKLDGSTHLN IFFAKSSEEE LAGVATWPWD KEALMH LGG IVLNPSFYGM PGHTHTMIHE IGHSLGLYHV FRGISEIQSC SDPCMETEPS FETGDLC ND TNPAPKHKSC GDPGPGNDTC GFHSFFNTPY NNFMSYADDD CTDSFTPNQV ARMHCYLD L VYQGWQPSRK PAPVALAPQV LGHTTDSVTL EWFPPIDGHF FERELGSACH LCLEGRILV QYASNASSPM PCSPSGHWSP REAEGHPDVE QPCKSSVRTW SPNSAVNPHT VPPACPEPQG CYLELEFLY PLVPESLTIW VTFVSTDWDS SGAVNDIKLL AVSGKNISLG PQNVFCDVPL T IRLWDVGE EVYGIQIYTL DEHLEIDAAM LTSTADTPLC LQCKPLKYKV VRDPPLQMDV AS ILHLNRK FVDMDLNLGS VYQYWVITIS GTEESEPSPA VTYIHGSGYC GDGIIQKDQG EQC DDMNKI NGDGCSLFCR QEVSFNCIDE PSRCYFHDGD GVCEEFEQKT SIKDCGVYTP QGFL DQWAS NASVSHQDQQ CPGWVIIGQP AASQVCRTKV IDLSEGISQH AWYPCTISYP YSQLA QTTF WLRAYFSQPM VAAAVIVHLV TDGTYYGDQK QETISVQLLD TKDQSHDLGL HVLSCR NNP LIIPVVHDLS QPFYHSQAVR VSFSSPLVAI SGVALRSFDN FDPVTLSSCQ RGETYSP AE QSCVHFACEK TDCPELAVEN ASLNCSSSDR YHGAQCTVSC RTGYVLQIRR DDELIKSQ T GPSVTVTCTE GKWNKQVACE PVDCSIPDHH QVYAASFSCP EGTTFGSQCS FQCRHPAQL KGNNSLLTCM EDGLWSFPEA LCELMCLAPP PVPNADLQTA RCRENKHKVG SFCKYKCKPG YHVPGSSRK SKKRAFKTQC TQDGSWQEGA CVPVTCDPPP PKFHGLYQCT NGFQFNSECR I KCEDSDAS QGLGSNVIHC RKDGTWNGSF HVCQEMQGQC SVPNELNSNL KLQCPDGYAI GS ECATSCL DHNSESIILP MNVTVRDIPH WLNPTRVERV VCTAGLKWYP HPALIHCVKG CEP FMGDNY CDAINNRAFC NYDGGDCCTS TVKTKKVTPF PMSCDLQGDC ACRDPQAQEH SRKD LRGYS HG GENBANK: GENBANK: Q13219 |
-Macromolecule #2: STC2
| Macromolecule | Name: STC2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: Extracellular Homo sapiens (human) / Cell: Extracellular |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MCAERLGQFM TLALVLATFD PARGTDATNP PEGPQDRSSQ QKGRLSLQNT AEIQHCLVNA GDVGCGVFE CFENNSCEIR GLHGICMTFL HNAGKFDAQG KSFIKDALKC KAHALRHRFG C ISRKCPAI REMVSQLQRE CYLKHDLCAA AQENTRVIVE MIHFKDLLLH ...String: MCAERLGQFM TLALVLATFD PARGTDATNP PEGPQDRSSQ QKGRLSLQNT AEIQHCLVNA GDVGCGVFE CFENNSCEIR GLHGICMTFL HNAGKFDAQG KSFIKDALKC KAHALRHRFG C ISRKCPAI REMVSQLQRE CYLKHDLCAA AQENTRVIVE MIHFKDLLLH EPYVDLVNLL LT CGEEVKE AITHSVQVQC EQNWGSLCSI LSFCTSAIQK PPTAPPERQP QVDRTKLSRA HHG EAGHHL PEPSSRETGR GAKGERGSKS HPNAHARGRV GGLGAQGPSG SSEWEDEQSE YSDI RR UniProtKB: Stanniocalcin-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: HEPES buffer |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)