+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1507 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of bacteriophage lambda procapsid | |||||||||
 Map data Map data | lambda prohead reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / phage / procapsid / prophage / lambda / icosahedral / cryoEM | |||||||||
| Biological species |  Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.5 Å | |||||||||
 Authors Authors | Lander GC / Evilevitch A / Jeembaeva M / Potter CS / Carragher B / Johnson JE | |||||||||
 Citation Citation |  Journal: Structure / Year: 2008 Journal: Structure / Year: 2008Title: Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Authors: Gabriel C Lander / Alex Evilevitch / Meerim Jeembaeva / Clinton S Potter / Bridget Carragher / John E Johnson /  Abstract: We report the cryo-EM structure of bacteriophage lambda and the mechanism for stabilizing the 20-A-thick capsid containing the dsDNA genome. The crystal structure of the HK97 bacteriophage capsid ...We report the cryo-EM structure of bacteriophage lambda and the mechanism for stabilizing the 20-A-thick capsid containing the dsDNA genome. The crystal structure of the HK97 bacteriophage capsid fits most of the T = 7 lambda particle density with only minor adjustment. A prominent surface feature at the 3-fold axes corresponds to the cementing protein gpD, which is necessary for stabilization of the capsid shell. Its position coincides with the location of the covalent cross-link formed in the docked HK97 crystal structure, suggesting an evolutionary replacement of this gene product in lambda by autocatalytic chemistry in HK97. The crystal structure of the trimeric gpD, in which the 14 N-terminal residues required for capsid binding are disordered, fits precisely into the corresponding EM density. The N-terminal residues of gpD are well ordered in the cryo-EM density, adding a strand to a beta-sheet formed by the capsid proteins and explaining the mechanism of particle stabilization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1507.map.gz emd_1507.map.gz | 19.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1507-v30.xml emd-1507-v30.xml emd-1507.xml emd-1507.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1507.gif 1507.gif | 77.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1507 http://ftp.pdbj.org/pub/emdb/structures/EMD-1507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1507 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1507.map.gz / Format: CCP4 / Size: 105.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1507.map.gz / Format: CCP4 / Size: 105.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | lambda prohead reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
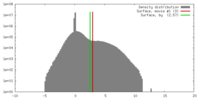
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : lambda procapsid
| Entire | Name: lambda procapsid |
|---|---|
| Components |
|
-Supramolecule #1000: lambda procapsid
| Supramolecule | Name: lambda procapsid / type: sample / ID: 1000 Details: particles were present in a preparation of a 37.7 kbp packaging lambda mutant Oligomeric state: icosahedral / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 16 MDa / Theoretical: 16 MDa |
-Supramolecule #1: Enterobacteria phage lambda
| Supramolecule | Name: Enterobacteria phage lambda / type: virus / ID: 1 / Name.synonym: capsid, gpE / NCBI-ID: 10710 / Sci species name: Enterobacteria phage lambda / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes / Syn species name: capsid, gpE |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Experimental: 16 MDa / Theoretical: 16 MDa |
| Virus shell | Shell ID: 1 / Name: gpE / Diameter: 500 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10mM MgSO4, 50mM Tris-HCl |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 77 K / Instrument: OTHER / Details: Vitrification instrument: FEI Vitrobot / Method: double blot for 7 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 77 K / Max: 78 K / Average: 78 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 135,000 times magnification |
| Date | Feb 16, 2007 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Number real images: 2683 / Average electron dose: 19 e/Å2 / Camera length: 61.4 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side-entry cryostage / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | Protocol: rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)