[English] 日本語
 Yorodumi
Yorodumi- EMDB-14913: Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conforma... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | ||||||||||||||||||
 Map data Map data | Cryo-EM map of the WT KdpFABC complex, in the E1_ATPearly conformation, at 3.5 A resolution, sharpened at -132 A2. Data obtained in the presence under turnover conditions | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | P-type ATPase / superfamily of K+ transporters (SKT) / potassium uptake system / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationP-type K+ transporter / P-type potassium transmembrane transporter activity / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / monoatomic cation transmembrane transport / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / ATP hydrolysis activity ...P-type K+ transporter / P-type potassium transmembrane transporter activity / potassium:proton antiporter complex / potassium ion-transporting ATPase complex / monoatomic cation transmembrane transport / potassium ion binding / potassium ion transmembrane transport / potassium ion transport / magnesium ion binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||
 Authors Authors | Hielkema L / Stock C / Silberberg JM / Corey RA / Rheinberger J / Wunnicke D / Dubach VRA / Stansfeld PJ / Haenelt I / Paulino C | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Inhibited KdpFABC transitions into an E1 off-cycle state. Authors: Jakob M Silberberg / Charlott Stock / Lisa Hielkema / Robin A Corey / Jan Rheinberger / Dorith Wunnicke / Victor R A Dubach / Phillip J Stansfeld / Inga Hänelt / Cristina Paulino /    Abstract: KdpFABC is a high-affinity prokaryotic K uptake system that forms a functional chimera between a channel-like subunit (KdpA) and a P-type ATPase (KdpB). At high K levels, KdpFABC needs to be ...KdpFABC is a high-affinity prokaryotic K uptake system that forms a functional chimera between a channel-like subunit (KdpA) and a P-type ATPase (KdpB). At high K levels, KdpFABC needs to be inhibited to prevent excessive K accumulation to the point of toxicity. This is achieved by a phosphorylation of the serine residue in the TGES motif in the A domain of the pump subunit KdpB (KdpB). Here, we explore the structural basis of inhibition by KdpB phosphorylation by determining the conformational landscape of KdpFABC under inhibiting and non-inhibiting conditions. Under turnover conditions, we identified a new inhibited KdpFABC state that we termed E1P tight, which is not part of the canonical Post-Albers transport cycle of P-type ATPases. It likely represents the biochemically described stalled E1P state adopted by KdpFABC upon KdpB phosphorylation. The E1P tight state exhibits a compact fold of the three cytoplasmic domains and is likely adopted when the transition from high-energy E1P states to E2P states is unsuccessful. This study represents a structural characterization of a biologically relevant off-cycle state in the P-type ATPase family and supports the emerging discussion of P-type ATPase regulation by such states. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14913.map.gz emd_14913.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14913-v30.xml emd-14913-v30.xml emd-14913.xml emd-14913.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14913_fsc.xml emd_14913_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14913.png emd_14913.png | 32.1 KB | ||
| Masks |  emd_14913_msk_1.map emd_14913_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14913.cif.gz emd-14913.cif.gz | 7.7 KB | ||
| Others |  emd_14913_half_map_1.map.gz emd_14913_half_map_1.map.gz emd_14913_half_map_2.map.gz emd_14913_half_map_2.map.gz | 40.8 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14913 http://ftp.pdbj.org/pub/emdb/structures/EMD-14913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14913 | HTTPS FTP |
-Related structure data
| Related structure data |  7zrgMC  7zrdC  7zreC  7zrhC  7zriC  7zrjC  7zrkC  7zrlC  7zrmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14913.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14913.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the WT KdpFABC complex, in the E1_ATPearly conformation, at 3.5 A resolution, sharpened at -132 A2. Data obtained in the presence under turnover conditions | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||
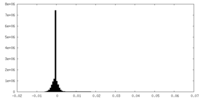
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14913_msk_1.map emd_14913_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
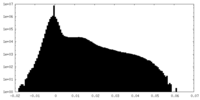
| Density Histograms |
-Half map: Half map 1 used during refinement and for...
| File | emd_14913_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 used during refinement and for FSC gold-standard resolution calculation of the WT KdpFABC complex in the E1_ATPearly conformation under turnover conditions | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 used during refinement and for...
| File | emd_14913_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 used during refinement and for FSC gold-standard resolution calculation of the WT KdpFABC complex in the E1_ATPearly conformation under turnover conditions | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KdpFABC
| Entire | Name: KdpFABC |
|---|---|
| Components |
|
-Supramolecule #1: KdpFABC
| Supramolecule | Name: KdpFABC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 157 KDa |
-Macromolecule #1: Potassium-transporting ATPase potassium-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase potassium-binding subunit type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.218613 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGLN MLGLAVLFFM LLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWQSYS GETTLSYFSQ MAGLTVQNFL SAASGIAVIF ALIRAFTRQS M STLGNAWV ...String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGLN MLGLAVLFFM LLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWQSYS GETTLSYFSQ MAGLTVQNFL SAASGIAVIF ALIRAFTRQS M STLGNAWV DLLRITLWVL VPVALLIALF FIQQGALQNF LPYQAVNTVE GAQQLLPMGP VASQEAIKML GTNGGGFFNA NS SHPFENP TALTNFVQML AIFLIPTALC FAFGEVMGDR RQGRMLLWAM SVIFVICVGV VMWAEVQGNP HLLALGTDSS INM EGKESR FGVLVSSLFA VVTTAASCGA VIAMHDSFTA LGGMVPMWLM QIGEVVFGGV GSGLYGMMLF VLLAVFIAGL MIGR TPEYL GKKIDVREMK LTALAILVTP TLVLMGAALA MMTDAGRSAM LNPGPHGFSE VLYAVSSAAN NNGSAFAGLS ANSPF WNCL LAFCMFVGRF GVIIPVMAIA GSLVSKKSQA ASSGTLPTHG PLFVGLLIGT VLLVGALTFI PALALGPVAE YLS UniProtKB: Potassium-transporting ATPase potassium-binding subunit |
-Macromolecule #2: Potassium-transporting ATPase KdpC subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpC subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.281035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGLRPALST FIFLLLITGG VYPLLTTVLG QWWFPWQANG SLIREGDTVR GSALIGQNFT GNGYFHGRPS ATAEMPYNPQ ASGGSNLAV SNPELDKLIA ARVAALRAAN PDASASVPVE LVTASASGLD NNITPQAAAW QIPRVAKARN LSVEQLTQLI A KYSQQPLV KYIGQPVVNI VELNLALDKL DE UniProtKB: Potassium-transporting ATPase KdpC subunit |
-Macromolecule #3: Potassium-transporting ATPase KdpF subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpF subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.853463 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAGVITGVL LVFLLLGYLV YALINAE UniProtKB: Potassium-transporting ATPase KdpF subunit |
-Macromolecule #4: Potassium-transporting ATPase ATP-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase ATP-binding subunit / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type K+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.347844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAISG WLWITVLFAN FAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD KVPADQLRKG DIVLVEAGDI IPCDGEVIEG GASVDESAIT G E(SEP)APVIRE ...String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAISG WLWITVLFAN FAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD KVPADQLRKG DIVLVEAGDI IPCDGEVIEG GASVDESAIT G E(SEP)APVIRE SGGDFASVTG GTRILSDWLV IECSVNPGET FLDRMIAMVE GAQRRKTPNE IALTILLIAL TIVFLLAT A TLWPFSAWGG NAVSVTVLVA LLVCLIPTTI GGLLSAIGVA GMSRMLGANV IATSGRAVEA AGDVDVLLLD KTGTITLGN RQASEFIPAQ GVDEKTLADA AQLASLADET PEGRSIVILA KQRFNLRERD VQSLHATFVP FTAQSRMSGI NIDNRMIRKG SVDAIRRHV EANGGHFPTD VDQKVDQVAR QGATPLVVVE GSRVLGVIAL KDIVKGGIKE RFAQLRKMGI KTVMITGDNR L TAAAIAAE AGVDDFLAEA TPEAKLALIR QYQAEGRLVA MTGDGTNDAP ALAQADVAVA MNSGTQAAKE AGNMVDLDSN PT KLIEVVH IGKQMLMTRG SLTTFSIAND VAKYFAIIPA AFAATYPQLN ALNIMCLHSP DSAILSAVIF NALIIVFLIP LAL KGVSYK PLTASAMLRR NLWIYGLGGL LVPFIGIKVI DLLLTVCGLV UniProtKB: Potassium-transporting ATPase ATP-binding subunit |
-Macromolecule #5: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 5 / Number of copies: 7 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #6: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 6 / Number of copies: 2 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #7: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM Tris-HCl pH 8, 10 mM MgCl2, 10 mM NaCl and 0.0125% DDM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Details: at 5 mA |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 3 / Number real images: 17938 / Average exposure time: 9.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


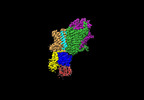












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)