[English] 日本語
 Yorodumi
Yorodumi- EMDB-14347: Cryo-EM map of the WT KdpFABC complex in the E2-P conformation, s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
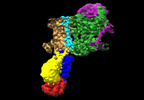
| Title | Cryo-EM map of the WT KdpFABC complex in the E2-P conformation, stabilised with the inhibitor orthovanadate | ||||||||||||||||||
 Map data Map data | Cryo-EM map of the WT KdpFABC complex, in the E2-P tight conformation, at 7.4 A resolution, sharpened at -195 A2. Data obtained in the presence of 2mM orthovanadate and 1mM KCl. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | P-type ATPase / superfamily of K+ transporters (SKT) / potassium uptake system / MEMBRANE PROTEIN | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
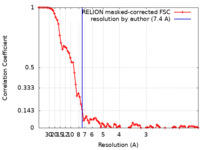
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | ||||||||||||||||||
 Authors Authors | Hielkema L / Stock C / Silberberg JM / Corey RA / Wunnicke D / Stansfeld PJ / Haenelt I / Paulino C | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Inhibited KdpFABC transitions into an E1 off-cycle state. Authors: Jakob M Silberberg / Charlott Stock / Lisa Hielkema / Robin A Corey / Jan Rheinberger / Dorith Wunnicke / Victor R A Dubach / Phillip J Stansfeld / Inga Hänelt / Cristina Paulino /    Abstract: KdpFABC is a high-affinity prokaryotic K uptake system that forms a functional chimera between a channel-like subunit (KdpA) and a P-type ATPase (KdpB). At high K levels, KdpFABC needs to be ...KdpFABC is a high-affinity prokaryotic K uptake system that forms a functional chimera between a channel-like subunit (KdpA) and a P-type ATPase (KdpB). At high K levels, KdpFABC needs to be inhibited to prevent excessive K accumulation to the point of toxicity. This is achieved by a phosphorylation of the serine residue in the TGES motif in the A domain of the pump subunit KdpB (KdpB). Here, we explore the structural basis of inhibition by KdpB phosphorylation by determining the conformational landscape of KdpFABC under inhibiting and non-inhibiting conditions. Under turnover conditions, we identified a new inhibited KdpFABC state that we termed E1P tight, which is not part of the canonical Post-Albers transport cycle of P-type ATPases. It likely represents the biochemically described stalled E1P state adopted by KdpFABC upon KdpB phosphorylation. The E1P tight state exhibits a compact fold of the three cytoplasmic domains and is likely adopted when the transition from high-energy E1P states to E2P states is unsuccessful. This study represents a structural characterization of a biologically relevant off-cycle state in the P-type ATPase family and supports the emerging discussion of P-type ATPase regulation by such states. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14347.map.gz emd_14347.map.gz | 49.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14347-v30.xml emd-14347-v30.xml emd-14347.xml emd-14347.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14347_fsc.xml emd_14347_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14347.png emd_14347.png | 58.5 KB | ||
| Masks |  emd_14347_msk_1.map emd_14347_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14347.cif.gz emd-14347.cif.gz | 6.6 KB | ||
| Others |  emd_14347_half_map_1.map.gz emd_14347_half_map_1.map.gz emd_14347_half_map_2.map.gz emd_14347_half_map_2.map.gz | 40.9 MB 40.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14347 http://ftp.pdbj.org/pub/emdb/structures/EMD-14347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14347 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14347.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14347.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the WT KdpFABC complex, in the E2-P tight conformation, at 7.4 A resolution, sharpened at -195 A2. Data obtained in the presence of 2mM orthovanadate and 1mM KCl. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||
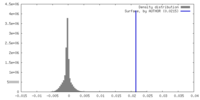
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14347_msk_1.map emd_14347_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
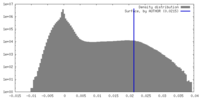
| Density Histograms |
-Half map: Half map 1 used during refinement and for...
| File | emd_14347_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 used during refinement and for FSC gold-standard resolution calculation of the WT KdpFABC complex in the presence of orthovanadate. | ||||||||||||
| Projections & Slices |
| ||||||||||||
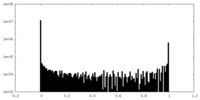
| Density Histograms |
-Half map: Half map 2 used during refinement and for...
| File | emd_14347_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 used during refinement and for FSC gold-standard resolution calculation of the WT KdpFABC complex in the presence of orthovanadate. | ||||||||||||
| Projections & Slices |
| ||||||||||||
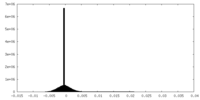
| Density Histograms |
- Sample components
Sample components
-Entire : KdpFABC
| Entire | Name: KdpFABC |
|---|---|
| Components |
|
-Supramolecule #1: KdpFABC
| Supramolecule | Name: KdpFABC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 157 KDa |
-Macromolecule #1: Potassium-transporting ATPase potassium-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase potassium-binding subunit type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGL NMLGLAVLFF MLLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWQSYS G ETTLSYFS QMAGLTVQNF LSAASGIAVI FALIRAFTRQ SMSTLGNAWV ...String: MAAQGFLLIA TFLLVLMVLA RPLGSGLARL INDIPLPGTT GVERVLFRAL GVSDREMNWK QYLCAILGL NMLGLAVLFF MLLGQHYLPL NPQQLPGLSW DLALNTAVSF VTNTNWQSYS G ETTLSYFS QMAGLTVQNF LSAASGIAVI FALIRAFTRQ SMSTLGNAWV DLLRITLWVL VP VALLIAL FFIQQGALQN FLPYQAVNTV EGAQQLLPMG PVASQEAIKM LGTNGGGFFN ANS SHPFEN PTALTNFVQM LAIFLIPTAL CFAFGEVMGD RRQGRMLLWA MSVIFVICVG VVMW AEVQG NPHLLALGTD SSINMEGKES RFGVLVSSLF AVVTTAASCG AVIAMHDSFT ALGGM VPMW LMQIGEVVFG GVGSGLYGMM LFVLLAVFIA GLMIGRTPEY LGKKIDVREM KLTALA ILV TPTLVLMGAA LAMMTDAGRS AMLNPGPHGF SEVLYAVSSA ANNNGSAFAG LSANSPF WN CLLAFCMFVG RFGVIIPVMA IAGSLVSKKS QAASSGTLPT HGPLFVGLLI GTVLLVGA L TFIPALALGP VAEYLS |
-Macromolecule #2: Potassium-transporting ATPase KdpC subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpC subunit / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSGLRPALST FIFLLLITGG VYPLLTTVLG QWWFPWQANG SLIREGDTVR GSALIGQNFT GNGYFHGRP SATAEMPYNP QASGGSNLAV SNPELDKLIA ARVAALRAAN PDASASVPVE L VTASASGL DNNITPQAAA WQIPRVAKAR NLSVEQLTQL IAKYSQQPLV KYIGQPVVNI VE LNLALDK LDE |
-Macromolecule #3: Potassium-transporting ATPase KdpF subunit
| Macromolecule | Name: Potassium-transporting ATPase KdpF subunit / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSAGVITGVL LVFLLLGYLV YALINAE |
-Macromolecule #4: Potassium-transporting ATPase ATP-binding subunit
| Macromolecule | Name: Potassium-transporting ATPase ATP-binding subunit / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO / EC number: P-type K+ transporter |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAIS GWLWITVLFA NFAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD K VPADQLRK GDIVLVEAGD IIPCDGEVIE GGASVDESAI TGE(SEP)APVIRE ...String: MSRKQLALFE PTLVVQALKE AVKKLNPQAQ WRNPVMFIVW IGSLLTTCIS IAMASGAMPG NALFSAAIS GWLWITVLFA NFAEALAEGR SKAQANSLKG VKKTAFARKL REPKYGAAAD K VPADQLRK GDIVLVEAGD IIPCDGEVIE GGASVDESAI TGE(SEP)APVIRE SGGDFASVTG GT RILSDWL VIECSVNPGE TFLDRMIAMV EGAQRRKTPN EIALTILLIA LTIVFLLATA TLW PFSAWG GNAVSVTVLV ALLVCLIPTT IGGLLSAIGV AGMSRMLGAN VIATSGRAVE AAGD VDVLL LDKTGTITLG NRQASEFIPA QGVDEKTLAD AAQLASLADE TPEGRSIVIL AKQRF NLRE RDVQSLHATF VPFTAQSRMS GINIDNRMIR KGSVDAIRRH VEANGGHFPT DVDQKV DQV ARQGATPLVV VEGSRVLGVI ALKDIVKGGI KERFAQLRKM GIKTVMITGD NRLTAAA IA AEAGVDDFLA EATPEAKLAL IRQYQAEGRL VAMTGDGTND APALAQADVA VAMNSGTQ A AKEAGNMVDL DSNPTKLIEV VHIGKQMLMT RGSLTTFSIA NDVAKYFAII PAAFAATYP QLNALNIMCL HSPDSAILSA VIFNALIIVF LIPLALKGVS YKPLTASAML RRNLWIYGLG GLLVPFIGI KVIDLLLTVC GLV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM Tris-HCl pH 8, 10 mM MgCl2, 10 mM NaCl and 0.0125% DDM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Details: at 5 mA |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 2 / Number real images: 2487 / Average exposure time: 9.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)