+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of an ALYREF-exon junction complex hexamer | |||||||||
 Map data Map data | Locally sharpened ALYREF(55-183)-EJC-RNA map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription-exort complex / TREX / splicing / exon junction complex / EJC / RNA export / RNA binding proteins / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationexon-exon junction subcomplex mago-y14 / negative regulation of selenocysteine incorporation / regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / cellular response to selenite ion / transcription export complex / selenocysteine insertion sequence binding / C5-methylcytidine-containing RNA reader activity / exon-exon junction complex / regulation of translation at postsynapse, modulating synaptic transmission / Z-decay: degradation of maternal mRNAs by zygotically expressed factors ...exon-exon junction subcomplex mago-y14 / negative regulation of selenocysteine incorporation / regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / cellular response to selenite ion / transcription export complex / selenocysteine insertion sequence binding / C5-methylcytidine-containing RNA reader activity / exon-exon junction complex / regulation of translation at postsynapse, modulating synaptic transmission / Z-decay: degradation of maternal mRNAs by zygotically expressed factors / regulation of mRNA processing / negative regulation of excitatory postsynaptic potential / Deadenylation of mRNA / embryonic cranial skeleton morphogenesis / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA / poly(A) binding / mRNA 3'-end processing / M-decay: degradation of maternal mRNAs by maternally stored factors / U2-type catalytic step 1 spliceosome / Transport of Mature mRNA Derived from an Intronless Transcript / RNA export from nucleus / Transport of Mature mRNA derived from an Intron-Containing Transcript / RNA Polymerase II Transcription Termination / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of alternative mRNA splicing, via spliceosome / exploration behavior / associative learning / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ribonucleoprotein complex binding / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / cellular response to brain-derived neurotrophic factor stimulus / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / positive regulation of translation / mRNA splicing, via spliceosome / ISG15 antiviral mechanism / Regulation of expression of SLITs and ROBOs / RNA stem-loop binding / mRNA processing / rRNA processing / osteoblast differentiation / regulation of translation / outer membrane-bounded periplasmic space / RNA helicase activity / postsynapse / negative regulation of translation / nuclear speck / RNA helicase / neuronal cell body / mRNA binding / dendrite / nucleolus / glutamatergic synapse / ATP hydrolysis activity / RNA binding / extracellular exosome / nucleoplasm / ATP binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
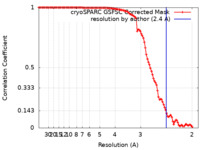
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Pacheco-Fiallos FB / Vorlaender MK / Plaschka C | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: mRNA recognition and packaging by the human transcription-export complex. Authors: Belén Pacheco-Fiallos / Matthias K Vorländer / Daria Riabov-Bassat / Laura Fin / Francis J O'Reilly / Farja I Ayala / Ulla Schellhaas / Juri Rappsilber / Clemens Plaschka /    Abstract: Newly made mRNAs are processed and packaged into mature ribonucleoprotein complexes (mRNPs) and are recognized by the essential transcription-export complex (TREX) for nuclear export. However, the ...Newly made mRNAs are processed and packaged into mature ribonucleoprotein complexes (mRNPs) and are recognized by the essential transcription-export complex (TREX) for nuclear export. However, the mechanisms of mRNP recognition and three-dimensional mRNP organization are poorly understood. Here we report cryo-electron microscopy and tomography structures of reconstituted and endogenous human mRNPs bound to the 2-MDa TREX complex. We show that mRNPs are recognized through multivalent interactions between the TREX subunit ALYREF and mRNP-bound exon junction complexes. Exon junction complexes can multimerize through ALYREF, which suggests a mechanism for mRNP organization. Endogenous mRNPs form compact globules that are coated by multiple TREX complexes. These results reveal how TREX may simultaneously recognize, compact and protect mRNAs to promote their packaging for nuclear export. The organization of mRNP globules provides a framework to understand how mRNP architecture facilitates mRNA biogenesis and export. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14803.map.gz emd_14803.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14803-v30.xml emd-14803-v30.xml emd-14803.xml emd-14803.xml | 31.2 KB 31.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14803_fsc.xml emd_14803_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14803.png emd_14803.png | 170.9 KB | ||
| Masks |  emd_14803_msk_1.map emd_14803_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14803.cif.gz emd-14803.cif.gz | 8.1 KB | ||
| Others |  emd_14803_additional_1.map.gz emd_14803_additional_1.map.gz emd_14803_additional_2.map.gz emd_14803_additional_2.map.gz emd_14803_half_map_1.map.gz emd_14803_half_map_1.map.gz emd_14803_half_map_2.map.gz emd_14803_half_map_2.map.gz | 117.9 MB 62.1 MB 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14803 http://ftp.pdbj.org/pub/emdb/structures/EMD-14803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14803 | HTTPS FTP |
-Validation report
| Summary document |  emd_14803_validation.pdf.gz emd_14803_validation.pdf.gz | 811.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14803_full_validation.pdf.gz emd_14803_full_validation.pdf.gz | 810.9 KB | Display | |
| Data in XML |  emd_14803_validation.xml.gz emd_14803_validation.xml.gz | 18.8 KB | Display | |
| Data in CIF |  emd_14803_validation.cif.gz emd_14803_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14803 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14803 | HTTPS FTP |
-Related structure data
| Related structure data |  7znjMC  7znkC  7znlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14803.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14803.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally sharpened ALYREF(55-183)-EJC-RNA map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.945 Å | ||||||||||||||||||||||||||||||||||||
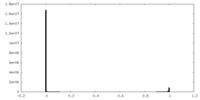
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14803_msk_1.map emd_14803_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
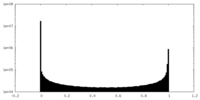
| Density Histograms |
-Additional map: Globally sharpened ALYREF(55-183)-EJC-RNA map
| File | emd_14803_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Globally sharpened ALYREF(55-183)-EJC-RNA map | ||||||||||||
| Projections & Slices |
| ||||||||||||
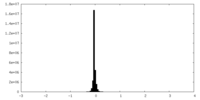
| Density Histograms |
-Additional map: Unsharpened ALYREF(55-183)-EJC-RNA map
| File | emd_14803_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened ALYREF(55-183)-EJC-RNA map | ||||||||||||
| Projections & Slices |
| ||||||||||||
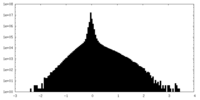
| Density Histograms |
-Half map: ALYREF(55-183)-EJC-RNA half map
| File | emd_14803_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ALYREF(55-183)-EJC-RNA half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ALYREF(55-183)-EJC-RNA half map
| File | emd_14803_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ALYREF(55-183)-EJC-RNA half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric complex between the TREX subunit ALYREF and the exon ju...
| Entire | Name: Hexameric complex between the TREX subunit ALYREF and the exon junction complex |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric complex between the TREX subunit ALYREF and the exon ju...
| Supramolecule | Name: Hexameric complex between the TREX subunit ALYREF and the exon junction complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Assembled with truncated MBP-tagged ALYREF (residues 55-183) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: Eukaryotic initiation factor 4A-III, N-terminally processed
| Macromolecule | Name: Eukaryotic initiation factor 4A-III, N-terminally processed type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.819367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKVEFETSE EVDVTPTFDT MGLREDLLRG IYAYGFEKPS AIQQRAIKQI IKGRDVIAQS QSGTGKTATF SISVLQCLDI QVRETQALI LAPTRELAVQ IQKGLLALGD YMNVQCHACI GGTNVGEDIR KLDYGQHVVA GTPGRVFDMI RRRSLRTRAI K MLVLDEAD ...String: MTKVEFETSE EVDVTPTFDT MGLREDLLRG IYAYGFEKPS AIQQRAIKQI IKGRDVIAQS QSGTGKTATF SISVLQCLDI QVRETQALI LAPTRELAVQ IQKGLLALGD YMNVQCHACI GGTNVGEDIR KLDYGQHVVA GTPGRVFDMI RRRSLRTRAI K MLVLDEAD EMLNKGFKEQ IYDVYRYLPP ATQVVLISAT LPHEILEMTN KFMTDPIRIL VKRDELTLEG IKQFFVAVER EE WKFDTLC DLYDTLTITQ AVIFCNTKRK VDWLTEKMRE ANFTVSSMHG DMPQKERESI MKEFRSGASR VLISTDVWAR GLD VPQVSL IINYDLPNNR ELYIHRIGRS GRYGRKGVAI NFVKNDDIRI LRDIEQYYST QIDEMP UniProtKB: Eukaryotic initiation factor 4A-III |
-Macromolecule #2: Protein mago nashi homolog
| Macromolecule | Name: Protein mago nashi homolog / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.189625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MESDFYLRYY VGHKGKFGHE FLEFEFRPDG KLRYANNSNY KNDVMIRKEA YVHKSVMEEL KRIIDDSEIT KEDDALWPPP DRVGRQELE IVIGDEHISF TTSKIGSLID VNQSKDPEGL RVFYYLVQDL KCLVFSLIGL HFKIKPI UniProtKB: Protein mago nashi homolog |
-Macromolecule #3: RNA-binding protein 8A
| Macromolecule | Name: RNA-binding protein 8A / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.370525 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPQRSVEGWI LFVTGVHEEA TEEDIHDKFA EYGEIKNIHL NLDRRTGYLK GYTLVEYETY KEAQAAMEGL NGQDLMGQPI SVDWCFVRG PP UniProtKB: RNA-binding protein 8A |
-Macromolecule #4: Maltose/maltodextrin-binding periplasmic protein,THO complex subunit 4
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,THO complex subunit 4 type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.598855 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQLSHHHHHH HHHHSSGMKI EEGKLVIWIN GDKGYNGLAE VGKKFEKDTG IKVTVEHPDK LEEKFPQVAA TGDGPDIIFW AHDRFGGYA QSGLLAEITP DKAFQDKLYP FTWDAVRYNG KLIAYPIAVE ALSLIYNKDL LPNPPKTWEE IPALDKELKA K GKSALMFN ...String: MQLSHHHHHH HHHHSSGMKI EEGKLVIWIN GDKGYNGLAE VGKKFEKDTG IKVTVEHPDK LEEKFPQVAA TGDGPDIIFW AHDRFGGYA QSGLLAEITP DKAFQDKLYP FTWDAVRYNG KLIAYPIAVE ALSLIYNKDL LPNPPKTWEE IPALDKELKA K GKSALMFN LQEPYFTWPL IAADGGYAFK YENGKYDIKD VGVDNAGAKA GLTFLVDLIK NKHMNADTDY SIAEAAFNKG ET AMTINGP WAWSNIDTSK VNYGVTVLPT FKGQPSKPFV GVLSAGINAA SPNKELAKEF LENYLLTDEG LEAVNKDKPL GAV ALKSYE EELAKDPRIA ATMENAQKGE IMPNIPQMSA FWYAVRTAVI NAASGRQTVD EALKDAQTSS GLEVLFQGPG SSGI RNRPA IARGAAGGGG RNRPAPYSRP KQLPDKWQHD LFDSGFGGGA GVETGGKLLV SNLDFGVSDA DIQELFAEFG TLKKA AVHY DRSGRSLGTA DVHFERKADA LKAMKQYNGV PLDGRPMNIQ LVT UniProtKB: Maltose/maltodextrin-binding periplasmic protein, THO complex subunit 4 |
-Macromolecule #5: RNA
| Macromolecule | Name: RNA / type: rna / ID: 5 / Number of copies: 6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.792037 KDa |
| Sequence | String: UUUUUU |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 7 / Number of copies: 6 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.7 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)