[English] 日本語
 Yorodumi
Yorodumi- EMDB-14518: Chimaera of AP2 with FCHO2 linker domain, N1-N2 enriched population -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chimaera of AP2 with FCHO2 linker domain, N1-N2 enriched population | |||||||||
 Map data Map data | N1-N2 enriched substructure of chimaera of AP2 with FCHO2 linker domain | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Kane Dickson V / Qu K / Owen DJ / Briggs JA / Zaccai NR | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P-dependent switch. Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / ...Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / Sally R Gray / Kun Qu / Philip R Evans / Marco Fritzsche / Filip Sroubek / Stefan Höning / John A G Briggs / Bernard T Kelly / David J Owen / Linton M Traub /      Abstract: Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane- ...Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane-localized Fer/Cip4 homology domain-only proteins (FCHO). Here, live-cell enhanced total internal reflection fluorescence-structured illumination microscopy shows that FCHO marks sites of clathrin-coated pit (CCP) initiation, which mature into uniform-sized CCPs comprising a central patch of AP2 and clathrin corralled by an FCHO/Epidermal growth factor potential receptor substrate number 15 (Eps15) ring. We dissect the network of interactions between the FCHO interdomain linker and AP2, which concentrates, orients, tethers, and partially destabilizes closed AP2 at the plasma membrane. AP2's subsequent membrane deposition drives its opening, which triggers FCHO displacement through steric competition with phosphatidylinositol 4,5-bisphosphate, clathrin, cargo, and CME accessory factors. FCHO can now relocate toward a CCP's outer edge to engage and activate further AP2s to drive CCP growth/maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14518.map.gz emd_14518.map.gz | 11.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14518-v30.xml emd-14518-v30.xml emd-14518.xml emd-14518.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14518.png emd_14518.png | 83.6 KB | ||
| Others |  emd_14518_half_map_1.map.gz emd_14518_half_map_1.map.gz emd_14518_half_map_2.map.gz emd_14518_half_map_2.map.gz | 139.3 MB 139.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14518 http://ftp.pdbj.org/pub/emdb/structures/EMD-14518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14518 | HTTPS FTP |
-Validation report
| Summary document |  emd_14518_validation.pdf.gz emd_14518_validation.pdf.gz | 670 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14518_full_validation.pdf.gz emd_14518_full_validation.pdf.gz | 669.5 KB | Display | |
| Data in XML |  emd_14518_validation.xml.gz emd_14518_validation.xml.gz | 14.5 KB | Display | |
| Data in CIF |  emd_14518_validation.cif.gz emd_14518_validation.cif.gz | 17.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14518 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14518 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14518 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14518 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14518.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14518.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | N1-N2 enriched substructure of chimaera of AP2 with FCHO2 linker domain | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.652 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_14518_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
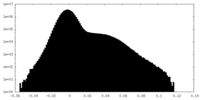
| Density Histograms |
-Half map: #2
| File | emd_14518_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AP2 core complex
| Entire | Name: AP2 core complex |
|---|---|
| Components |
|
-Supramolecule #1: AP2 core complex
| Supramolecule | Name: AP2 core complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Ternary complex of AP2 core expressed as part of a chimaera with FCHO2 linker (not modelled) |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 203 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 50% HKT buffer (10mM Hepes, 10mM Tris 120mM potassium acetate pH 7.2) and 50% Core buffer (10mM Tris, 250mM NaCl, pH 8) |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 28392 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)