[English] 日本語
 Yorodumi
Yorodumi- EMDB-14525: AP2 adaptor protein recruited on the membrane in the presence of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | AP2 adaptor protein recruited on the membrane in the presence of FCHO2 linker | |||||||||
 Map data Map data | Relion_postprocess generated map, low-pass filtered according to local resolution, sharpened with global B factor -1100 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adaptins / endocytosis / clathrin-mediated / FCHO2 / clathrin adaptor | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
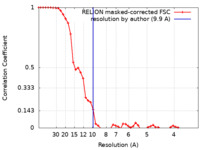
| Method | electron tomography / cryo EM / Resolution: 9.9 Å | |||||||||
 Authors Authors | Kovtun O / Kaufman JGG / Owen DJ / Briggs JAG | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P-dependent switch. Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / ...Authors: Nathan R Zaccai / Zuzana Kadlecova / Veronica Kane Dickson / Kseniya Korobchevskaya / Jan Kamenicky / Oleksiy Kovtun / Perunthottathu K Umasankar / Antoni G Wrobel / Jonathan G G Kaufman / Sally R Gray / Kun Qu / Philip R Evans / Marco Fritzsche / Filip Sroubek / Stefan Höning / John A G Briggs / Bernard T Kelly / David J Owen / Linton M Traub /      Abstract: Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane- ...Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane-localized Fer/Cip4 homology domain-only proteins (FCHO). Here, live-cell enhanced total internal reflection fluorescence-structured illumination microscopy shows that FCHO marks sites of clathrin-coated pit (CCP) initiation, which mature into uniform-sized CCPs comprising a central patch of AP2 and clathrin corralled by an FCHO/Epidermal growth factor potential receptor substrate number 15 (Eps15) ring. We dissect the network of interactions between the FCHO interdomain linker and AP2, which concentrates, orients, tethers, and partially destabilizes closed AP2 at the plasma membrane. AP2's subsequent membrane deposition drives its opening, which triggers FCHO displacement through steric competition with phosphatidylinositol 4,5-bisphosphate, clathrin, cargo, and CME accessory factors. FCHO can now relocate toward a CCP's outer edge to engage and activate further AP2s to drive CCP growth/maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14525.map.gz emd_14525.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14525-v30.xml emd-14525-v30.xml emd-14525.xml emd-14525.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14525_fsc.xml emd_14525_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_14525.png emd_14525.png | 51.3 KB | ||
| Masks |  emd_14525_msk_1.map emd_14525_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14525.cif.gz emd-14525.cif.gz | 6.9 KB | ||
| Others |  emd_14525_half_map_1.map.gz emd_14525_half_map_1.map.gz emd_14525_half_map_2.map.gz emd_14525_half_map_2.map.gz | 7.6 MB 7.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14525 http://ftp.pdbj.org/pub/emdb/structures/EMD-14525 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14525 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14525 | HTTPS FTP |
-Validation report
| Summary document |  emd_14525_validation.pdf.gz emd_14525_validation.pdf.gz | 367.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14525_full_validation.pdf.gz emd_14525_full_validation.pdf.gz | 367.3 KB | Display | |
| Data in XML |  emd_14525_validation.xml.gz emd_14525_validation.xml.gz | 10.3 KB | Display | |
| Data in CIF |  emd_14525_validation.cif.gz emd_14525_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14525 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14525 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14525 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14525 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14525.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14525.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion_postprocess generated map, low-pass filtered according to local resolution, sharpened with global B factor -1100 | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.701 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14525_msk_1.map emd_14525_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened halfmap 1
| File | emd_14525_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened halfmap 2
| File | emd_14525_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Clathrin adaptor protein AP2 recruited on the membrane in the pre...
| Entire | Name: Clathrin adaptor protein AP2 recruited on the membrane in the presence of interdomain linker of FCHO2 |
|---|---|
| Components |
|
-Supramolecule #1: Clathrin adaptor protein AP2 recruited on the membrane in the pre...
| Supramolecule | Name: Clathrin adaptor protein AP2 recruited on the membrane in the presence of interdomain linker of FCHO2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: AP-2 complex subunit alpha-2
| Macromolecule | Name: AP-2 complex subunit alpha-2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MPAVSKGDGM RGLAVFISD I RNCKSKEA EI KRINKEL ANI RSKFKG DKAL DGYSK KKYVC KLLF IFLLGH DID FGHMEAV NL LSSNRYTE K QIGYLFISV LVNSNSELIR LINNAIKND L ASRNPTFM GL ALHCIAN VGS REMAEA FAGE IPKIL ...String: MPAVSKGDGM RGLAVFISD I RNCKSKEA EI KRINKEL ANI RSKFKG DKAL DGYSK KKYVC KLLF IFLLGH DID FGHMEAV NL LSSNRYTE K QIGYLFISV LVNSNSELIR LINNAIKND L ASRNPTFM GL ALHCIAN VGS REMAEA FAGE IPKIL VAGDT MDSV KQSAAL CLL RLYRTSP DL VPMGDWTS R VVHLLNDQH LGVVTAATSL ITTLAQKNP E EFKTSVSL AV SRLSRIV TSA STDLQD YTYY FVPAP WLSVK LLRL LQCYPP PED PAVRGRL TE CLETILNK A QEPPKSKKV QHSNAKNAVL FEAISLIIH H DSEPNLLV RA CNQLGQF LQH RETNLR YLAL ESMCT LASSE FSHE AVKTHI ETV INALKTE RD VSVRQRAV D LLYAMCDRS NAQQIVAEML SYLETADYS I REEIVLKV AI LAEKYAV DYT WYVDTI LNLI RIAGD YVSEE VWYR VIQIVI NRD DVQGYAA KT VFEALQAP A CHENLVKVG GYILGEFGNL IAGDPRSSP L IQFNLLHS KF HLCSVPT RAL LLSTYI KFVN LFPEV KATIQ DVLR SDSQLK NAD VELQQRA VE YLRLSTVA S TDILATVLE EMPPFPERES SILAKLKKK K GGSGLVPR |
-Macromolecule #2: AP-2 complex subunit beta
| Macromolecule | Name: AP-2 complex subunit beta / type: protein_or_peptide / ID: 2 / Details: N-terminal His-tag / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHMTD SKYFTTNKK G EIFELKAE LN NEKKEKR KEA VKKVIA AMTV GKDVS SLFPD VVNC MQTDNL ELK KLVYLYL MN YAKSQPDM A IMAVNSFVK DCEDPNPLIR ALAVRTMGC I RVDKITEY LC EPLRKCL KDE DPYVRK TAAV CVAKL ...String: MHHHHHHMTD SKYFTTNKK G EIFELKAE LN NEKKEKR KEA VKKVIA AMTV GKDVS SLFPD VVNC MQTDNL ELK KLVYLYL MN YAKSQPDM A IMAVNSFVK DCEDPNPLIR ALAVRTMGC I RVDKITEY LC EPLRKCL KDE DPYVRK TAAV CVAKL HDINA QMVE DQGFLD SLR DLIADSN PM VVANAVAA L SEISESHPN SNLLDLNPQN INKLLTALN E CTEWGQIF IL DCLSNYN PKD DREAQS ICER VTPRL SHANS AVVL SAVKVL MKF LELLPKD SD YYNMLLKK L APPLVTLLS GEPEVQYVAL RNINLIVQK R PEILKQEI KV FFVKYND PIY VKLEKL DIMI RLASQ ANIAQ VLAE LKEYAT EVD VDFVRKA VR AIGRCAIK V EQSAERCVS TLLDLIQTKV NYVVQEAIV V IRDIFRKY PN KYESIIA TLC ENLDSL DEPD ARAAM IWIVG EYAE RIDNAD ELL ESFLEGF HD ESTQVQLT L LTAIVKLFL KKPSETQELV QQVLSLATQ D SDNPDLRD RG YIYWRLL STD PVTAKE VVLS EKPLI SEETD LIEP TLLDEL ICH IGSLASV YH KPPNAFVE G SHGIHRKHL PIHHGSTDAG DSPVGTTTA T NLEQPQVI PS QGDLLGD LLN LDLGPP VNVP QVSSM QMGAV DLLG GGLDSL VGQ SFIPSSV PA TFAPSPTP A VVSSGLNDL FELSTGIGMA PGGYVAPKA V WLPAVKAK GL EISGTFT HRQ GHIYME MNFT NKALQ HMTDF AIQF NKNSFG VIP STPLAIH TP LMPNQSID V SLPLNTLGP VMKMEPLNNL QVAVKNNID V FYFSCLIP LN VLFVEDG KME RQVFLA TWKD IPNEN ELQFQ IKEC HLNADT VSS KLQNNNV YT IAKRNVEG Q DMLYQSLKL TNGIWILAEL RIQPGNPNY T LSLKCRAP EV SQYIYQV YDS ILKN |
-Macromolecule #3: AP-2 complex subunit mu
| Macromolecule | Name: AP-2 complex subunit mu / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MIGGLFIYNH KGEVLISRV Y RDDIGRNA VD AFRVNVI HAR QQVRSP VTNI ARTSF FHVKR SNIW LAAVTK QNV NAAMVFE FL YKMCDVMA A YFGKISEEN IKNNFVLIYE LLDEILDFG Y PQNSETGA LK TFITQQG IKS QHQTKE EQSQ ITSQV ...String: MIGGLFIYNH KGEVLISRV Y RDDIGRNA VD AFRVNVI HAR QQVRSP VTNI ARTSF FHVKR SNIW LAAVTK QNV NAAMVFE FL YKMCDVMA A YFGKISEEN IKNNFVLIYE LLDEILDFG Y PQNSETGA LK TFITQQG IKS QHQTKE EQSQ ITSQV TGQIG WRRE GIKYRR NEL FLDVLES VN LLMSPQGQ V LSAHVSGRV VMKSYLSGMP ECKFGMNDK I VIEKQGKG TA DETSKSM EQK LISEED LGKQ SIAID DCTFH QCVR LSKFDS ERS ISFIPPD GE FELMRYRT T KDIILPFRV IPLVREVGRT KLEVKVVIK S NFKPSLLA QK IEVRIPT PLN TSGVQV ICMK GKAKY KASEN AIVW KIKRMA GMK ESQISAE IE LLPTNDKK K WARPPISMN FEVPFAPSGL KVRYLKVFE P KLNYSDHD VI KWVRYIG RSG IYETRC |
-Macromolecule #4: AP-2 complex subunit sigma
| Macromolecule | Name: AP-2 complex subunit sigma / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MIRFILIQNR AGKTRLAKW Y MQFDDDEK QK LIEEVHA VVT VRDAKH TNFV EFRNF KIIYR RYAG LYFCIC VDV NDNNLAY LE AIHNFVEV L NEYFHNVCE LDLVFNFYKV YTVVDEMFL A GEIRETSQ TK VLKQLLM LQS LE |
-Macromolecule #5: FCHO2: F-BAR domain only protein 2 interdomain linker
| Macromolecule | Name: FCHO2: F-BAR domain only protein 2 interdomain linker / type: protein_or_peptide / ID: 5 / Details: Interdomain linker / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: daesvecpda dslnipdvd e egysikpe tn qndtken hfy sssdsd sede epkky rieik pmhp nnshht mas ldelkvs ig nitlspai s rhspvqmnr nlsneeltks kpsappnek g tsdllawd pl fgpslds sss ss |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: BBI / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.17 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)