[English] 日本語
 Yorodumi
Yorodumi- EMDB-14013: Structure of recombinant human gamma-Tubulin Ring Complex 12-spok... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14013 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of recombinant human gamma-Tubulin Ring Complex 12-spoked assembly intermediate (spokes 3-14, substoichiometric spokes 1-2) | |||||||||
 Map data Map data | 12-spoked assembly intermediate of the gamma-Tubulin Ring Complex (spokes 3-14, substoichiometric 1-2) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Assembly / Intermediate / Complex / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / polar microtubule / interphase microtubule organizing center / gamma-tubulin complex / gamma-tubulin ring complex / mitotic spindle microtubule / meiotic spindle organization / microtubule nucleation ...microtubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / polar microtubule / interphase microtubule organizing center / gamma-tubulin complex / gamma-tubulin ring complex / mitotic spindle microtubule / meiotic spindle organization / microtubule nucleation / gamma-tubulin binding / non-motile cilium / pericentriolar material / cell leading edge / microtubule organizing center / mitotic sister chromatid segregation / single fertilization / mitotic spindle assembly / cytoplasmic microtubule / spindle assembly / cytoplasmic microtubule organization / centriole / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / condensed nuclear chromosome / mitotic spindle organization / meiotic cell cycle / brain development / recycling endosome / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / neuron migration / spindle pole / apical part of cell / Regulation of PLK1 Activity at G2/M Transition / mitotic cell cycle / microtubule cytoskeleton / protein-containing complex assembly / microtubule binding / microtubule / neuron projection / ciliary basal body / cilium / centrosome / GTP binding / structural molecule activity / nucleoplasm / identical protein binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.7 Å | |||||||||
 Authors Authors | Zupa E / Pfeffer S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Modular assembly of the principal microtubule nucleator γ-TuRC. Authors: Martin Würtz / Erik Zupa / Enrico S Atorino / Annett Neuner / Anna Böhler / Ariani S Rahadian / Bram J A Vermeulen / Giulia Tonon / Sebastian Eustermann / Elmar Schiebel / Stefan Pfeffer /  Abstract: The gamma-tubulin ring complex (γ-TuRC) is the principal microtubule nucleation template in vertebrates. Recent cryo-EM reconstructions visualized the intricate quaternary structure of the γ-TuRC, ...The gamma-tubulin ring complex (γ-TuRC) is the principal microtubule nucleation template in vertebrates. Recent cryo-EM reconstructions visualized the intricate quaternary structure of the γ-TuRC, containing more than thirty subunits, raising fundamental questions about γ-TuRC assembly and the role of actin as an integral part of the complex. Here, we reveal the structural mechanism underlying modular γ-TuRC assembly and identify a functional role of actin in microtubule nucleation. During γ-TuRC assembly, a GCP6-stabilized core comprising GCP2-3-4-5-4-6 is expanded by stepwise recruitment, selective stabilization and conformational locking of four pre-formed GCP2-GCP3 units. Formation of the lumenal bridge specifies incorporation of the terminal GCP2-GCP3 unit and thereby leads to closure of the γ-TuRC ring in a left-handed spiral configuration. Actin incorporation into the complex is not relevant for γ-TuRC assembly and structural integrity, but determines γ-TuRC geometry and is required for efficient microtubule nucleation and mitotic chromosome alignment in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14013.map.gz emd_14013.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14013-v30.xml emd-14013-v30.xml emd-14013.xml emd-14013.xml | 28.7 KB 28.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14013.png emd_14013.png | 114.5 KB | ||
| Filedesc metadata |  emd-14013.cif.gz emd-14013.cif.gz | 10.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14013 http://ftp.pdbj.org/pub/emdb/structures/EMD-14013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14013 | HTTPS FTP |
-Related structure data
| Related structure data |  7qj8MC  7qj0C  7qj1C  7qj2C  7qj3C  7qj4C  7qj5C  7qj6C  7qj7C  7qj9C  7qjaC  7qjbC  7qjcC  7qjdC  7qjeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14013.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14013.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 12-spoked assembly intermediate of the gamma-Tubulin Ring Complex (spokes 3-14, substoichiometric 1-2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
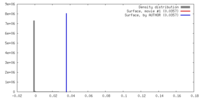
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Recombinant human Gamma-Tubulin Ring Complex
+Supramolecule #1: Recombinant human Gamma-Tubulin Ring Complex
+Macromolecule #1: Gamma-tubulin complex component 5
+Macromolecule #2: Tubulin gamma-1 chain
+Macromolecule #3: actin, cytoplasmic 1
+Macromolecule #4: Mitotic-spindle organizing protein 1
+Macromolecule #5: Gamma-tubulin complex component 3
+Macromolecule #6: Gamma-tubulin complex component 2
+Macromolecule #7: Gamma-tubulin complex component 4
+Macromolecule #8: Gamma-tubulin complex component 6
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6127 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||||||||||||||
| Output model |  PDB-7qj8: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















 Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths)
Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths)




