[English] 日本語
 Yorodumi
Yorodumi- EMDB-14006: Structure of the recombinant human gamma-Tubulin Ring Complex 6-s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14006 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the recombinant human gamma-Tubulin Ring Complex 6-spoked assembly intermediate (spokes 7-12, homogeneous dataset) | |||||||||
 Map data Map data | 6-spoked assembly intermediate of the gamma-Tubulin Ring Complex, homogeneous dataset, (spokes 7-12) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Assembly / Intermediate / Complex / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / polar microtubule / interphase microtubule organizing center / gamma-tubulin complex / gamma-tubulin ring complex / mitotic spindle microtubule / meiotic spindle organization / microtubule nucleation ...microtubule nucleation by interphase microtubule organizing center / gamma-tubulin complex localization / microtubule nucleator activity / polar microtubule / interphase microtubule organizing center / gamma-tubulin complex / gamma-tubulin ring complex / mitotic spindle microtubule / meiotic spindle organization / microtubule nucleation / gamma-tubulin binding / non-motile cilium / pericentriolar material / cell leading edge / microtubule organizing center / mitotic sister chromatid segregation / mitotic spindle assembly / single fertilization / cytoplasmic microtubule / spindle assembly / cytoplasmic microtubule organization / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / centriole / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / condensed nuclear chromosome / mitotic spindle organization / meiotic cell cycle / recycling endosome / brain development / structural constituent of cytoskeleton / microtubule cytoskeleton organization / spindle / neuron migration / apical part of cell / spindle pole / Regulation of PLK1 Activity at G2/M Transition / mitotic cell cycle / microtubule cytoskeleton / protein-containing complex assembly / microtubule binding / microtubule / neuron projection / cilium / ciliary basal body / centrosome / GTP binding / structural molecule activity / nucleoplasm / identical protein binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Zupa E / Pfeffer S | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Modular assembly of the principal microtubule nucleator γ-TuRC. Authors: Martin Würtz / Erik Zupa / Enrico S Atorino / Annett Neuner / Anna Böhler / Ariani S Rahadian / Bram J A Vermeulen / Giulia Tonon / Sebastian Eustermann / Elmar Schiebel / Stefan Pfeffer /  Abstract: The gamma-tubulin ring complex (γ-TuRC) is the principal microtubule nucleation template in vertebrates. Recent cryo-EM reconstructions visualized the intricate quaternary structure of the γ-TuRC, ...The gamma-tubulin ring complex (γ-TuRC) is the principal microtubule nucleation template in vertebrates. Recent cryo-EM reconstructions visualized the intricate quaternary structure of the γ-TuRC, containing more than thirty subunits, raising fundamental questions about γ-TuRC assembly and the role of actin as an integral part of the complex. Here, we reveal the structural mechanism underlying modular γ-TuRC assembly and identify a functional role of actin in microtubule nucleation. During γ-TuRC assembly, a GCP6-stabilized core comprising GCP2-3-4-5-4-6 is expanded by stepwise recruitment, selective stabilization and conformational locking of four pre-formed GCP2-GCP3 units. Formation of the lumenal bridge specifies incorporation of the terminal GCP2-GCP3 unit and thereby leads to closure of the γ-TuRC ring in a left-handed spiral configuration. Actin incorporation into the complex is not relevant for γ-TuRC assembly and structural integrity, but determines γ-TuRC geometry and is required for efficient microtubule nucleation and mitotic chromosome alignment in vivo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14006.map.gz emd_14006.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14006-v30.xml emd-14006-v30.xml emd-14006.xml emd-14006.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14006.png emd_14006.png | 75.3 KB | ||
| Filedesc metadata |  emd-14006.cif.gz emd-14006.cif.gz | 9.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14006 http://ftp.pdbj.org/pub/emdb/structures/EMD-14006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14006 | HTTPS FTP |
-Related structure data
| Related structure data |  7qj1MC  7qj0C  7qj2C  7qj3C  7qj4C  7qj5C  7qj6C  7qj7C  7qj8C  7qj9C  7qjaC  7qjbC  7qjcC  7qjdC  7qjeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14006.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14006.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 6-spoked assembly intermediate of the gamma-Tubulin Ring Complex, homogeneous dataset, (spokes 7-12) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
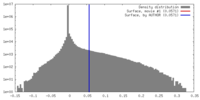
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Recombinant human Gamma-Tubulin Ring Complex
| Entire | Name: Recombinant human Gamma-Tubulin Ring Complex |
|---|---|
| Components |
|
-Supramolecule #1: Recombinant human Gamma-Tubulin Ring Complex
| Supramolecule | Name: Recombinant human Gamma-Tubulin Ring Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.0 MDa |
-Macromolecule #1: Gamma-tubulin complex component 5
| Macromolecule | Name: Gamma-tubulin complex component 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 118.467547 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MARHGPPWSR LDAQQERDVR ELVRGVAGLQ DEADPNFQLA LNFAWSNFRF HRFLDVNSHK IEKTIEGIYE KFVIHSDLSK AASWKRLTE EFLNAPLPSI KEIKTDAHYS ILSLLLCLSD SPSNSSYVET PRNKEVEKKD DFDWGKYLME DEEMDIGPYM D TPNWSEES ...String: MARHGPPWSR LDAQQERDVR ELVRGVAGLQ DEADPNFQLA LNFAWSNFRF HRFLDVNSHK IEKTIEGIYE KFVIHSDLSK AASWKRLTE EFLNAPLPSI KEIKTDAHYS ILSLLLCLSD SPSNSSYVET PRNKEVEKKD DFDWGKYLME DEEMDIGPYM D TPNWSEES EEENDQQPLS REDSGIQVDR TPLEEQDQNR KLDPCISWKD EPDDRSWLEH HVVHQYWTAR PSQFPHSLHL HS NLAAVWD QHLYSSDPLY VPDDRVLVTE TQVIRETLWL LSGVKKLFIF QLIDGKVTVR NNIIVTHLTH SCLRSVLEQI AAY GQVVFR LQEFIDEVMG HSSESMLPGS GSVPKKSTEA PFRTYQAFMW ALYKYFISFK EELAEIEKCI INNDTTITLA IVVD KLAPR LSQLKVLHKV FSTGVAEVPP DTRNVVRASH LLNTLYKAIL EYDNVGEASE QTVSLLFSLW VETVRPYLQT VDEWI VHGH LWDGAREFII QRNKNVPVNH RDFWYATYTL YSVSEKTENE EKMSDNASAS SGSDQGPSSR QHTMVSFLKP VLKQII MAG KSMQLLKNLQ CAESTTCQAG ARDAERKSLY TLFLESVQSR LRHGEDSTPQ VLTEQQATKE NLMKMQSIAE SHLELDD VH DPLLAINFAR MYLEQSDFHE KFAGGDVCVD RSSESVTCQT FELTLRSCLY PHIDKQYLDC CGNLMQTLKK DYRLVEYL Q AMRNFFLMEG GDTMYDFYTS IFDKIREKET WQNVSFLNVQ LQEAVGQRYP EDSSRLSISF ENVDTAKKKL PVHILDGLT LSYKVPWPVD IVISLECQKI YNQVFLLLLQ IKWAKYSLDV LLFGELVSTA EKPRLKEGLI HEQDTVAQFG PQKEPVRQQI HRMFLLRVK LMHFVNSLHN YIMTRILHST GLEFQHQVEE AKDLDQLIKI HYRYLSTIHD RCLLREKVSF VKEAIMKVLN L ALMFADGW QAGLGTWRME SIEKMESDFK NCHMFLVTIL NKAVCRGSFP HLESLALSLM AGMEQS UniProtKB: Gamma-tubulin complex component 5 |
-Macromolecule #2: Mitotic-spindle organizing protein 1
| Macromolecule | Name: Mitotic-spindle organizing protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.485724 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MASSSGAGAA AAAAAANLNA VRETMDVLLE ISRILNTGLD METLSICVRL CEQGINPEAL SSVIKELRKA TEALKAAENM TS UniProtKB: Mitotic-spindle organizing protein 1 |
-Macromolecule #3: Gamma-tubulin complex component 3
| Macromolecule | Name: Gamma-tubulin complex component 3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 103.710102 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MATPDQKSPN VLLQNLCCRI LGRSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELIRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRRQPS KVSSYATLFA QALPRDAHST PYYYARPQTL PLSYQDRSAQ SAQSSGSVGS S GISSIGLC ...String: MATPDQKSPN VLLQNLCCRI LGRSEADVAQ QFQYAVRVIG SNFAPTVERD EFLVAEKIKK ELIRQRREAD AALFSELHRK LHSQGVLKN KWSILYLLLS LSEDPRRQPS KVSSYATLFA QALPRDAHST PYYYARPQTL PLSYQDRSAQ SAQSSGSVGS S GISSIGLC ALSGPAPAPQ SLLPGQSNQA PGVGDCLRQQ LGSRLAWTLT ANQPSSQATT SKGVPSAVSR NMTRSRREGD TG GTMEITE AALVRDILYV FQGIDGKNIK MNNTENCYKV EGKANLSRSL RDTAVRLSEL GWLHNKIRRY TDQRSLDRSF GLV GQSFCA ALHQELREYY RLLSVLHSQL QLEDDQGVNL GLESSLTLRR LLVWTYDPKI RLKTLAALVD HCQGRKGGEL ASAV HAYTK TGDPYMRSLV QHILSLVSHP VLSFLYRWIY DGELEDTYHE FFVASDPTVK TDRLWHDKYT LRKSMIPSFM TMDQS RKVL LIGKSINFLH QVCHDQTPTT KMIAVTKSAE SPQDAADLFT DLENAFQGKI DAAYFETSKY LLDVLNKKYS LLDHMQ AMR RYLLLGQGDF IRHLMDLLKP ELVRPATTLY QHNLTGILET AVRATNAQFD SPEILRRLDV RLLEVSPGDT GWDVFSL DY HVDGPIATVF TRECMSHYLR VFNFLWRAKR MEYILTDIRK GHMCNAKLLR NMPEFSGVLH QCHILASEMV HFIHQMQY Y ITFEVLECSW DELWNKVQQA QDLDHIIAAH EVFLDTIISR CLLDSDSRAL LNQLRAVFDQ IIELQNAQDA IYRAALEEL QRRLQFEEKK KQREIEGQWG VTAAEEEEEN KRIGEFKESI PKMCSQLRIL THFYQGIVQQ FLVLLTTSSD ESLRFLSFRL DFNEHYKAR EPRLRVSLGT RGRRSSHT UniProtKB: Gamma-tubulin complex component 3 |
-Macromolecule #4: Gamma-tubulin complex component 2
| Macromolecule | Name: Gamma-tubulin complex component 2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 102.666953 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLT EDKETLQYLQ QNAKERAELA AAAVGSSTTS INVPAAASKI SMQELEELRK QLGSVATGST LQQSLELKRK M LRDKQNKK ...String: MSEFRIHHDV NELLSLLRVH GGDGAEVYID LLQKNRTPYV TTTVSAHSAK VKIAEFSRTP EDFLKKYDEL KSKNTRNLDP LVYLLSKLT EDKETLQYLQ QNAKERAELA AAAVGSSTTS INVPAAASKI SMQELEELRK QLGSVATGST LQQSLELKRK M LRDKQNKK NSGQHLPIFP AWVYERPALI GDFLIGAGIS TDTALPIGTL PLASQESAVV EDLLYVLVGV DGRYVSAQPL AG RQSRTFL VDPNLDLSIR ELVHRILPVA ASYSAVTRFI EEKSSFEYGQ VNHALAAAMR TLVKEHLILV SQLEQLHRQG LLS LQKLWF YIQPAMRTMD ILASLATSVD KGECLGGSTL SLLHDRSFSY TGDSQAQELC LYLTKAASAP YFEVLEKWIY RGII HDPYS EFMVEEHELR KERIQEDYND KYWDQRYTIV QQQIPSFLQK MADKILSTGK YLNVVRECGH DVTCPVAKEI IYTLK ERAY VEQIEKAFNY ASKVLLDFLM EEKELVAHLR SIKRYFLMDQ GDFFVHFMDL AEEELRKPVE DITPPRLEAL LELALR MST ANTDPFKDDL KIDLMPHDLI TQLLRVLAIE TKQEKAMAHA DPTELALSGL EAFSFDYIVK WPLSLIINRK ALTRYQM LF RHMFYCKHVE RQLCSVWISN KTAKQHSLHS AQWFAGAFTL RQRMLNFVQN IQYYMMFEVM EPTWHILEKN LKSASNID D VLGHHTGFLD TCLKDCMLTN PELLKVFSKL MSVCVMFTNC MQKFTQSMKL DGELGGQTLE HSTVLGLPAG AEERARKEL ARKHLAEHAD TVQLVSGFEA TINKFDKNFS AHLLDLLARL SIYSTSDCEH GMASVISRLD FNGFYTERLE RLSAERSQKA TPQVPVLRG PPAPAPRVAV TAQ UniProtKB: Gamma-tubulin complex component 2 |
-Macromolecule #5: Gamma-tubulin complex component 4
| Macromolecule | Name: Gamma-tubulin complex component 4 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.179969 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MIHELLLALS GYPGSIFTWN KRSGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLGDPHLSI SHVNYFLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV ...String: MIHELLLALS GYPGSIFTWN KRSGLQVSQD FPFLHPSETS VLNRLCRLGT DYIRFTEFIE QYTGHVQQQD HHPSQQGQGG LHGIYLRAF CTGLDSVLQP YRQALLDLEQ EFLGDPHLSI SHVNYFLDQF QLLFPSVMVV VEQIKSQKIH GCQILETVYK H SCGGLPPV RSALEKILAV CHGVMYKQLS AWMLHGLLLD QHEEFFIKQG PSSGNVSAQP EEDEEDLGIG GLTGKQLREL QD LRLIEEE NMLAPSLKQF SLRVEILPSY IPVRVAEKIL FVGESVQMFE NQNVNLTRKG SILKNQEDTF AAELHRLKQQ PLF SLVDFE QVVDRIRSTV AEHLWKLMVE ESDLLGQLKI IKDFYLLGRG ELFQAFIDTA QHMLKTPPTA VTEHDVNVAF QQSA HKVLL DDDNLLPLLH LTIEYHGKEH KADATQAREG PSRETSPREA PASGWAALGL SYKVQWPLHI LFTPAVLEKY NVVFK YLLS VRRVQAELQH CWALQMQRKH LKSNQTDAIK WRLRNHMAFL VDNLQYYLQV DVLESQFSQL LHQINSTRDF ESIRLA HDH FLSNLLAQSF ILLKPVFHCL NEILDLCHSF CSLVSQNLGP LDERGAAQLS ILVKGFSRQS SLLFKILSSV RNHQINS DL AQLLLRLDYN KYYTQAGGTL GSFGM UniProtKB: Gamma-tubulin complex component 4 |
-Macromolecule #6: Gamma-tubulin complex component 6
| Macromolecule | Name: Gamma-tubulin complex component 6 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200.733641 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MASITQLFDD LCEALLPAAK THLGQRSVNR KRAKRSLKKV AYNALFTNLF QDETQQLQPD MSKLPARNKI LMLSFDLRVG GLGPKADRL EELVEELEAA PCCPLLEVGS VLDLLVQLAG SGPPQVLPRK RDYFLNNKHV GRNVPYSGYD CDDLSVFEMD V QSLISREE ...String: MASITQLFDD LCEALLPAAK THLGQRSVNR KRAKRSLKKV AYNALFTNLF QDETQQLQPD MSKLPARNKI LMLSFDLRVG GLGPKADRL EELVEELEAA PCCPLLEVGS VLDLLVQLAG SGPPQVLPRK RDYFLNNKHV GRNVPYSGYD CDDLSVFEMD V QSLISREE CLCHSMIQET LQVMEAAPGT GLPTVGLFSF GDPCGDRFER DTRVSLFGAL VHSRTYDMDV RLGLPPVPDN AD LSGLAIK VPPSVDQWED EGFQSASNLT PDSQSEPSVT PDVDLWEAAL TYEASKRRCW ERVGCPPGHR EEPYLTEAGR DAF DKFCRL HQGELQLLAG GVLQAPQPVL VKECELVKDV LNVLIGVVSA TFSLCQPAQA FVVKRGVHVS GASPESISSL LSEV AEYGT CYTRLSHFSL QPVLDSLYSK GLVFQAFTSG LRRYLQYYRA CVLSTPPTLS LLTIGFLFKK LGRQLRYLAE LCGVG AVLP GTCGGGPRAA FPTGVKLLSY LYQEALHNCS NEHYPVLLSL LKTSCEPYTR FIHDWVYSGV FRDAYGEFMI QVNHEY LSF RDKLYWTHGY VLISKEVEDC VPVFLKHIAH DIYVCGKTIN LLKLCCPRHY LCWSDVPVPR ISVIFSLEEL KEIEKDC AV YVGRMERVAR HSSVSKEEKE LRMEIAKQEL IAHAREAASR VLSALSDRQM SERMALDARK REQFQRLKEQ FVKDQERR Q AARQEELDDD FSYARELRDR ERRLKSLEEE LERKARQALV DHYSKLSAEA ARREQKALWR IQRHRLESAR LRFLLEDEK HIQEMLKAVS EAHQPQEPPD VLLSVHPQVT SPGPEHPEGG QGCDSGSAEQ HSPAWDGWNR PGLLTPQPLK PLAVGAGGRG LQQAEGARP FSDSLSIGDF LPVGPGAEPS VQTGMVPLLE VALQTINLDL PPSAPGEAPA AASTQPSRPQ EYDFSTVLRP A VATSPAPG PLQAAECSLG SSGLQLWEDS CGKMDACGSA SRETLLPSHP PRRAALEEGS SQPTERLFGQ VSGGGLPTGD YA SEIAPTR PRWNTHGHVS DASIRVGENV SDVAPTQPRW NTHGHVSNAS ISLGESVSDV APTRPRWNIH GHVSNASIRV GEN VSDVAP TRPRWNTHGH VSNASIRVGE NVSDVAPTRP RWNTHGHVSD ASISLGESVS DMAPARPRWN THGHVSDASI SLGE SVSDM APTRPRWNTH GHVSDTSIRV GENVSDVAPI RSRCNTHGHV SDASISLGEP VSDVVSTRPR WNTHVPIPPP HMVLG ALSP EAEPNTPRPQ QSPPGHTSQS ALSLGAQSTV LDCGPRLPVE VGPSLSSPSS GCGEGSISVG ENVSDVAPTQ PWWPNT PGD SVSEELGPGR SGDTEDLSPN WPLNSQEDTA AQSSPGRGEE AEASAAEAQG GEQAYLAGLA GQYHLERYPD SYESMSE PP IAHLLRPVLP RAFAFPVDPQ VQSAADETAV QLSELLTLPV LMKRSITAPL AAHISLVNKA AVDYFFVELH LEAHYEAL R HFLLMEDGEF AQSLSDLLFE KLGAGQTPGE LLNPLVLNSV LSKALQCSLH GDTPHASNLS LALKYLPEVF APNAPDVLS CLELRYKVDW PLNIVITEGC VSKYSGVFSF LLQLKLMMWA LKDVCFHLKR TALLSHMAGS VQFRQLQLFK HEMQHFVKVI QGYIANQIL HVTWCEFRAR LATVGDLEEI QRAHAEYLHK AVFRGLLTEK AAPVMNVIHS IFSLVLKFRS QLISQAWGPP G GPRGAEHP NFALMQQSYN TFKYYSHFLF KVVTKLVNRG YQPHLEDFLL RINFNNYYQD A UniProtKB: Gamma-tubulin complex component 6 |
-Macromolecule #7: Tubulin gamma-1 chain
| Macromolecule | Name: Tubulin gamma-1 chain / type: protein_or_peptide / ID: 7 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.22777 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT ...String: MPREIITLQL GQCGNQIGFE FWKQLCAEHG ISPEGIVEEF ATEGTDRKDV FFYQADDEHY IPRAVLLDLE PRVIHSILNS PYAKLYNPE NIYLSEHGGG AGNNWASGFS QGEKIHEDIF DIIDREADGS DSLEGFVLCH SIAGGTGSGL GSYLLERLND R YPKKLVQT YSVFPNQDEM SDVVVQPYNS LLTLKRLTQN ADCVVVLDNT ALNRIATDRL HIQNPSFSQI NQLVSTIMSA ST TTLRYPG YMNNDLIGLI ASLIPTPRLH FLMTGYTPLT TDQSVASVRK TTVLDVMRRL LQPKNVMVST GRDRQTNHCY IAI LNIIQG EVDPTQVHKS LQRIRERKLA NFIPWGPASI QVALSRKSPY LPSAHRVSGL MMANHTSISS LFERTCRQYD KLRK REAFL EQFRKEDMFK DNFDEMDTSR EIVQQLIDEY HAATRPDYIS WGTQEQ UniProtKB: Tubulin gamma-1 chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6127 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||||||||||||||
| Output model |  PDB-7qj1: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)