[English] 日本語
 Yorodumi
Yorodumi- EMDB-13993: In situ structure of myosin neck domain in skeletal sarcomere (ce... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13993 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of myosin neck domain in skeletal sarcomere (centered on regulatory light chain) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Skeletal muscle / Sarcomere / Cross-bridge / MOTOR PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / Smooth Muscle Contraction / muscle myosin complex / myosin filament / myosin complex / structural constituent of muscle / response to muscle activity / myofibril / cytoskeletal motor activity / skeletal muscle tissue development ...Striated Muscle Contraction / Smooth Muscle Contraction / muscle myosin complex / myosin filament / myosin complex / structural constituent of muscle / response to muscle activity / myofibril / cytoskeletal motor activity / skeletal muscle tissue development / muscle contraction / actin filament binding / double-stranded RNA binding / calmodulin binding / calcium ion binding / ATP binding Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
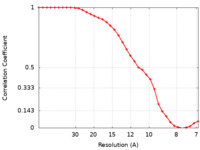
| Method | subtomogram averaging / cryo EM / Resolution: 9.0 Å | ||||||||||||||||||
 Authors Authors | Wang Z / Grange M | ||||||||||||||||||
| Funding support | European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Authors: Zhexin Wang / Michael Grange / Sabrina Pospich / Thorsten Wagner / Ay Lin Kho / Mathias Gautel / Stefan Raunser /   Abstract: In skeletal muscle, nebulin stabilizes and regulates the length of thin filaments, but the underlying mechanism remains nebulous. In this work, we used cryo-electron tomography and subtomogram ...In skeletal muscle, nebulin stabilizes and regulates the length of thin filaments, but the underlying mechanism remains nebulous. In this work, we used cryo-electron tomography and subtomogram averaging to reveal structures of native nebulin bound to thin filaments within intact sarcomeres. This in situ reconstruction provided high-resolution details of the interaction between nebulin and actin, demonstrating the stabilizing role of nebulin. Myosin bound to the thin filaments exhibited different conformations of the neck domain, highlighting its inherent structural variability in muscle. Unexpectedly, nebulin did not interact with myosin or tropomyosin, but it did interact with a troponin T linker through two potential binding motifs on nebulin, explaining its regulatory role. Our structures support the role of nebulin as a thin filament "molecular ruler" and provide a molecular basis for studying nemaline myopathies. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13993.map.gz emd_13993.map.gz | 153.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13993-v30.xml emd-13993-v30.xml emd-13993.xml emd-13993.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13993_fsc.xml emd_13993_fsc.xml | 3.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13993.png emd_13993.png | 27.5 KB | ||
| Masks |  emd_13993_msk_1.map emd_13993_msk_1.map | 2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13993.cif.gz emd-13993.cif.gz | 6.4 KB | ||
| Others |  emd_13993_half_map_1.map.gz emd_13993_half_map_1.map.gz emd_13993_half_map_2.map.gz emd_13993_half_map_2.map.gz | 1.4 MB 1.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13993 http://ftp.pdbj.org/pub/emdb/structures/EMD-13993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13993 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13993 | HTTPS FTP |
-Related structure data
| Related structure data |  7qioMC  7qimC  7qinC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13993.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13993.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13993_msk_1.map emd_13993_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13993_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13993_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse psoas muscle myofibrils
| Entire | Name: Mouse psoas muscle myofibrils |
|---|---|
| Components |
|
-Supramolecule #1: Mouse psoas muscle myofibrils
| Supramolecule | Name: Mouse psoas muscle myofibrils / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Myosin-4
| Macromolecule | Name: Myosin-4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 96.405609 KDa |
| Sequence | String: MSSDAEMAVF GEAAPYLRKS EKERIEAQNK PFDAKSSVFV VDAKESYVKA TVQSREGGKV TAKTEGGATV TVKDDQVFSM NPPKYDKIE DMAMMTHLHE PAVLYNLKER YAAWMIYTYS GLFCVTVNPY KWLPVYNPEV VAAYRGKKRQ EAPPHIFSIS D NAYQFMLT ...String: MSSDAEMAVF GEAAPYLRKS EKERIEAQNK PFDAKSSVFV VDAKESYVKA TVQSREGGKV TAKTEGGATV TVKDDQVFSM NPPKYDKIE DMAMMTHLHE PAVLYNLKER YAAWMIYTYS GLFCVTVNPY KWLPVYNPEV VAAYRGKKRQ EAPPHIFSIS D NAYQFMLT DRENQSILIT GESGAGKTVN TKRVIQYFAT IAVTGDKKKE EATSGKMQGT LEDQIISANP LLEAFGNAKT VR NDNSSRF GKFIRIHFGA TGKLASADIE TYLLEKSRVT FQLKAERSYH IFYQIMSNKK PELIEMLLIT TNPYDFAYVS QGE ITVPSI DDQEELMATD TAVDILGFSA DEKVAIYKLT GAVMHYGNMK FKQKQREEQA EPDGTEVADK AAYLTSLNSA DLLK ALCYP RVKVGNEYVT KGQTVQQVYN SVGALAKSMY EKMFLWMVTR INQQLDTKQP RQYFIGVLDI AGFEIFDFNT LEQLC INFT NEKLQQFFNH HMFVLEQEEY KKEGIDWEFI DFGMDLAACI ELIEKPMGIF SILEEECMFP KATDTSFKNK LYEQHL GKS NNFQKPKPAK GKAEAHFSLV HYAGTVDYNI IGWLDKNKDP LNETVVGLYQ KSGLKTLAFL FSGGQAAEAE GGGGKKG GK KKGSSFQTVS ALFRENLNKL MTNLKSTHPH FVRCLIPNET KTPGAMEHEL VLHQLRCNGV LEGIRICRKG FPSRILYA D FKQRYKVLNA SAIPEGQFID SKKASEKLLG SIDIDHTQYK FGHTKVFFKA GLLGTLEEMR DEKLAQLITR TQAVCRGYL MRVEFKKMME RRESIFCIQY NVRAFMNVKH WPWMKLYFKI KPLLKSAE UniProtKB: Myosin-4 |
-Macromolecule #2: Myosin light chain 1/3, skeletal muscle isoform
| Macromolecule | Name: Myosin light chain 1/3, skeletal muscle isoform / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.62049 KDa |
| Sequence | String: MAPKKDVKKP AAAPAPAPAP APAPAKPKEE KIDLSAIKIE FSKEQQEDFK EAFLLFDRTG ECKITLSQVG DVLRALGTNP TNAEVKKVL GNPSNEEMNA KKIEFEQFLP MMQAISNNKD QGGYEDFVEG LRVFDKEGNG TVMGAELRHV LATLGEKMKE E EVEALLAG QEDSNGCINY EAFVKHIMSV UniProtKB: Myosin light chain 1/3, skeletal muscle isoform |
-Macromolecule #3: Myosin regulatory light chain 2, skeletal muscle isoform
| Macromolecule | Name: Myosin regulatory light chain 2, skeletal muscle isoform type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.978445 KDa |
| Sequence | String: MAPKKAKRRA GAEGSSNVFS MFDQTQIQEF KEAFTVIDQN RDGIIDKEDL RDTFAAMGRL NVKNEELDAM MKEASGPINF TVFLTMFGE KLKGADPEDV ITGAFKVLDP EGKGTIKKQF LEELLTTQCD RFSQEEIKNM WAAFPPDVGG NVDYKNICYV I THGDAKDQ E UniProtKB: Myosin regulatory light chain 11 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 2.4 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.4 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 300 | ||||||||
| Output model |  PDB-7qio: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)