[English] 日本語
 Yorodumi
Yorodumi- EMDB-13831: Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13831 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | |||||||||
 Map data Map data | Cryo-EM map of Ty3 intasome targeting RNA Pol III transcribed gene | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription / RNA Pol III / Ty3 Retrotransposon / Intasome / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase III core binding / TFIIA-class transcription factor complex binding / DNA-templated transcription open complex formation / RNA polymerase III transcription regulatory region sequence-specific DNA binding / RNA polymerase III preinitiation complex assembly / transcription factor TFIIIB complex / TFIIIC-class transcription factor complex binding / RNA polymerase I general transcription initiation factor binding / RNA polymerase III type 3 promoter sequence-specific DNA binding / regulation of transcription by RNA polymerase III ...RNA polymerase III core binding / TFIIA-class transcription factor complex binding / DNA-templated transcription open complex formation / RNA polymerase III transcription regulatory region sequence-specific DNA binding / RNA polymerase III preinitiation complex assembly / transcription factor TFIIIB complex / TFIIIC-class transcription factor complex binding / RNA polymerase I general transcription initiation factor binding / RNA polymerase III type 3 promoter sequence-specific DNA binding / regulation of transcription by RNA polymerase III / RNA polymerase III general transcription initiation factor activity / transcription factor TFIIA complex / RNA polymerase I preinitiation complex assembly / transcription preinitiation complex / ribonuclease H / Hydrolases; Acting on peptide bonds (peptidases); Aspartic endopeptidases / RNA Polymerase III Transcription Initiation From Type 2 Promoter / DNA binding, bending / RNA polymerase II transcribes snRNA genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA polymerase II general transcription initiation factor activity / transcription factor TFIID complex / RNA Polymerase II Pre-transcription Events / RNA Polymerase I Promoter Escape / nucleolar large rRNA transcription by RNA polymerase I / Estrogen-dependent gene expression / transcription by RNA polymerase III / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / TBP-class protein binding / DNA-templated transcription initiation / DNA integration / RNA-directed DNA polymerase / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / disordered domain specific binding / DNA recombination / transcription regulator complex / DNA-binding transcription factor binding / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-directed DNA polymerase activity / viral translational frameshifting / negative regulation of DNA-templated transcription / chromatin binding / regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II / protein-containing complex / proteolysis / DNA binding / RNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
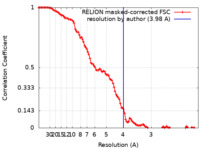
| Method | single particle reconstruction / cryo EM / Resolution: 3.98 Å | |||||||||
 Authors Authors | Abascal-Palacios G / Jochem L / Pla-Prats C / Beuron F / Vannini A | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes. Authors: Guillermo Abascal-Palacios / Laura Jochem / Carlos Pla-Prats / Fabienne Beuron / Alessandro Vannini /    Abstract: Retrotransposons are endogenous elements that have the ability to mobilise their DNA between different locations in the host genome. The Ty3 retrotransposon integrates with an exquisite specificity ...Retrotransposons are endogenous elements that have the ability to mobilise their DNA between different locations in the host genome. The Ty3 retrotransposon integrates with an exquisite specificity in a narrow window upstream of RNA Polymerase (Pol) III-transcribed genes, representing a paradigm for harmless targeted integration. Here we present the cryo-EM reconstruction at 4.0 Å of an active Ty3 strand transfer complex bound to TFIIIB transcription factor and a tRNA gene. The structure unravels the molecular mechanisms underlying Ty3 targeting specificity at Pol III-transcribed genes and sheds light into the architecture of retrotransposon machinery during integration. Ty3 intasome contacts a region of TBP, a subunit of TFIIIB, which is blocked by NC2 transcription regulator in RNA Pol II-transcribed genes. A newly-identified chromodomain on Ty3 integrase interacts with TFIIIB and the tRNA gene, defining with extreme precision the integration site position. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13831.map.gz emd_13831.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13831-v30.xml emd-13831-v30.xml emd-13831.xml emd-13831.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13831_fsc.xml emd_13831_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13831.png emd_13831.png | 123.3 KB | ||
| Filedesc metadata |  emd-13831.cif.gz emd-13831.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13831 http://ftp.pdbj.org/pub/emdb/structures/EMD-13831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13831 | HTTPS FTP |
-Related structure data
| Related structure data |  7q5bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13831.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13831.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Ty3 intasome targeting RNA Pol III transcribed gene | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
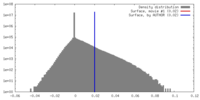
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Ty3 retrotransposon engaged with TFIIIB transcription factor and ...
+Supramolecule #1: Ty3 retrotransposon engaged with TFIIIB transcription factor and ...
+Supramolecule #2: DNA
+Supramolecule #3: TFIIIB transcription factor
+Macromolecule #1: DNA (56-MER)
+Macromolecule #2: DNA (31-MER)
+Macromolecule #3: DNA (34-MER)
+Macromolecule #4: DNA (9-MER)
+Macromolecule #9: DNA (19-MER)
+Macromolecule #5: Transposon Ty3-G Gag-Pol polyprotein
+Macromolecule #6: Transcription factor TFIIIB component B''
+Macromolecule #7: TATA-box-binding protein
+Macromolecule #8: Transcription factor IIIB 70 kDa subunit
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: Glow discharged at 15 mA | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||
| Details | This sample was monodisperse |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 4974 / Average exposure time: 7.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2437 / Average exposure time: 5.3 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.4 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: Correlation Coefficient |
|---|---|
| Output model |  PDB-7q5b: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)