+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | GA1 bacteriophage portal protein | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Bacteriophage / Portal protein / cryoEM / VIRAL PROTEIN | |||||||||||||||
| Function / homology | Portal protein Gp10 / Portal protein Gp10 superfamily / Phage Connector (GP10) / Head-tail connector (Portal protein) Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Bacillus phage GA-1 (virus) Bacillus phage GA-1 (virus) | |||||||||||||||
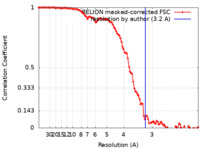
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Javed A / Villanueva H | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Viruses / Year: 2021 Journal: Viruses / Year: 2021Title: Cryo-EM Structures of Two Bacteriophage Portal Proteins Provide Insights for Antimicrobial Phage Engineering. Authors: Abid Javed / Hugo Villanueva / Shadikejiang Shataer / Sara Vasciaveo / Renos Savva / Elena V Orlova /  Abstract: Widespread antibiotic resistance has returned attention to bacteriophages as a means of managing bacterial pathogenesis. Synthetic biology approaches to engineer phages have demonstrated genomic ...Widespread antibiotic resistance has returned attention to bacteriophages as a means of managing bacterial pathogenesis. Synthetic biology approaches to engineer phages have demonstrated genomic editing to broaden natural host ranges, or to optimise microbicidal action. Gram positive pathogens cause serious pastoral animal and human infections that are especially lethal in newborns. Such pathogens are targeted by the obligate lytic phages of the and families. These phages have relatively small ~20 kb linear protein-capped genomes and their compact organisation, relatively few structural elements, and broad host range, are appealing from a phage-engineering standpoint. In this study, we focus on portal proteins, which are core elements for the assembly of such tailed phages. The structures of dodecameric portal complexes from phage GA1, which targets , and phage phiCPV4 that infects , were determined at resolutions of 3.3 Å and 2.9 Å, respectively. Both are found to closely resemble the related phi29 portal protein fold. However, the portal protein of phiCPV4 exhibits interesting differences in the clip domain. These structures provide new insights on structural diversity in portal proteins and will be essential for considerations in phage structural engineering. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13664.map.gz emd_13664.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13664-v30.xml emd-13664-v30.xml emd-13664.xml emd-13664.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13664_fsc.xml emd_13664_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13664.png emd_13664.png | 97.2 KB | ||
| Filedesc metadata |  emd-13664.cif.gz emd-13664.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13664 http://ftp.pdbj.org/pub/emdb/structures/EMD-13664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13664 | HTTPS FTP |
-Related structure data
| Related structure data |  7pv2MC  7pv4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13664.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13664.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : GA1 bacteriophage portal protein
| Entire | Name: GA1 bacteriophage portal protein |
|---|---|
| Components |
|
-Supramolecule #1: GA1 bacteriophage portal protein
| Supramolecule | Name: GA1 bacteriophage portal protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: A dodecamer complex made up of 12 subunits. |
|---|---|
| Source (natural) | Organism:  Bacillus phage GA-1 (virus) Bacillus phage GA-1 (virus) |
-Macromolecule #1: Head-tail connector (Portal protein)
| Macromolecule | Name: Head-tail connector (Portal protein) / type: protein_or_peptide / ID: 1 Details: In each subunit of the dodecamer, the DNA Tunnel loop residues (232 - 250) has not been modelled due to lack of EM density. Also missing in the pdb structure are 22 residues at the C- ...Details: In each subunit of the dodecamer, the DNA Tunnel loop residues (232 - 250) has not been modelled due to lack of EM density. Also missing in the pdb structure are 22 residues at the C-terminus and 7 residues at the N-terminus in each chain. Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillus phage GA-1 (virus) Bacillus phage GA-1 (virus) |
| Molecular weight | Theoretical: 35.307801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLEDGFSYKT IGEIQRRRGN LWFRTYQRYL FSLAYQMFEW QGLPKTVDPI FLEKQLHQRG FVAFYKDEMY GYLGVQGTLS GQINLYNQP NFYTASAPTY QKSFPLYWYD MGEDLNEKGQ GIVIYNNLER MPTLDILNLY AMNLAELKET IYVNQNAQKT P VIIKAGDN ...String: MLEDGFSYKT IGEIQRRRGN LWFRTYQRYL FSLAYQMFEW QGLPKTVDPI FLEKQLHQRG FVAFYKDEMY GYLGVQGTLS GQINLYNQP NFYTASAPTY QKSFPLYWYD MGEDLNEKGQ GIVIYNNLER MPTLDILNLY AMNLAELKET IYVNQNAQKT P VIIKAGDN DLFSMKQVYN KYEGNEPVIF AGKKFNTDDI EVLKTDAPYV ADKLTMLFKD QWNEAMTFLG LSNANTDKKE RL IQSEVES NNDQIQGSAN IYLAPRQEAC RLINEYYGLN VSVKLRKELV GNGELHNAIE GNSGMGNTV UniProtKB: Head-tail connector (Portal protein) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: 3ul of sample was applied on lacey carbon grids.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 95.0 K / Max: 98.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-12 / Number grids imaged: 1 / Number real images: 2249 / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)