+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13581 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human SLFN5 | |||||||||
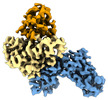
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleotide binding protein antiviral linked to tumorigenesis transcription regulation / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Lammens K / Metzner FJ | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Structural and biochemical characterization of human Schlafen 5. Authors: Felix J Metzner / Elisabeth Huber / Karl-Peter Hopfner / Katja Lammens /  Abstract: The Schlafen family belongs to the interferon-stimulated genes and its members are involved in cell cycle regulation, T cell quiescence, inhibition of viral replication, DNA-repair and tRNA ...The Schlafen family belongs to the interferon-stimulated genes and its members are involved in cell cycle regulation, T cell quiescence, inhibition of viral replication, DNA-repair and tRNA processing. Here, we present the cryo-EM structure of full-length human Schlafen 5 (SLFN5) and the high-resolution crystal structure of the highly conserved N-terminal core domain. We show that the core domain does not resemble an ATPase-like fold and neither binds nor hydrolyzes ATP. SLFN5 binds tRNA as well as single- and double-stranded DNA, suggesting a potential role in transcriptional regulation. Unlike rat Slfn13 or human SLFN11, human SLFN5 did not cleave tRNA. Based on the structure, we identified two residues in proximity to the zinc finger motif that decreased DNA binding when mutated. These results indicate that Schlafen proteins have divergent enzymatic functions and provide a structural platform for future biochemical and genetic studies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13581.map.gz emd_13581.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13581-v30.xml emd-13581-v30.xml emd-13581.xml emd-13581.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13581.png emd_13581.png | 152.4 KB | ||
| Filedesc metadata |  emd-13581.cif.gz emd-13581.cif.gz | 5.6 KB | ||
| Others |  emd_13581_half_map_1.map.gz emd_13581_half_map_1.map.gz emd_13581_half_map_2.map.gz emd_13581_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13581 http://ftp.pdbj.org/pub/emdb/structures/EMD-13581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13581 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13581 | HTTPS FTP |
-Validation report
| Summary document |  emd_13581_validation.pdf.gz emd_13581_validation.pdf.gz | 747.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13581_full_validation.pdf.gz emd_13581_full_validation.pdf.gz | 747.1 KB | Display | |
| Data in XML |  emd_13581_validation.xml.gz emd_13581_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_13581_validation.cif.gz emd_13581_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13581 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13581 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13581 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13581 | HTTPS FTP |
-Related structure data
| Related structure data |  7ppjMC  6rr9C  7q3zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13581.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13581.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_13581_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
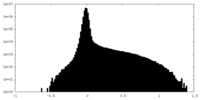
| Projections & Slices |
| ||||||||||||
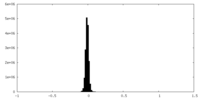
| Density Histograms |
-Half map: #1
| File | emd_13581_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
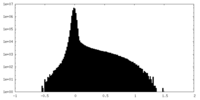
| Projections & Slices |
| ||||||||||||
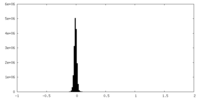
| Density Histograms |
- Sample components
Sample components
-Entire : full length human Schlafen 5
| Entire | Name: full length human Schlafen 5 |
|---|---|
| Components |
|
-Supramolecule #1: full length human Schlafen 5
| Supramolecule | Name: full length human Schlafen 5 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Schlafen family member 5
| Macromolecule | Name: Schlafen family member 5 / type: protein_or_peptide / ID: 1 Details: N-terminal 2 x Flag tag HRV 3C site flexible C-terminus Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104.420016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDDDDK GTDYKDDDDK LEVLFQGPMS LRIDVDTNFP ECVVDAGKVT LGTQQRQEMD PRLREKQNEI ILRAVCALLN SGGGIIKAE IENKGYNYER HGVGLDVPPI FRSHLDKMQK ENHFLIFVKS WNTEAGVPLA TLCSNLYHRE RTSTDVMDSQ E ALAFLKCR ...String: MADYKDDDDK GTDYKDDDDK LEVLFQGPMS LRIDVDTNFP ECVVDAGKVT LGTQQRQEMD PRLREKQNEI ILRAVCALLN SGGGIIKAE IENKGYNYER HGVGLDVPPI FRSHLDKMQK ENHFLIFVKS WNTEAGVPLA TLCSNLYHRE RTSTDVMDSQ E ALAFLKCR TQTPTNINVS NSLGPQAAQG SVQYEGNINV SAAALFDRKR LQYLEKLNLP ESTHVEFVMF STDVSHCVKD RL PKCVSAF ANTEGGYVFF GVHDETCQVI GCEKEKIDLT SLRASIDGCI KKLPVHHFCT QRPEIKYVLN FLEVHDKGAL RGY VCAIKV EKFCCAVFAK VPSSWQVKDN RVRQLPTREW TAWMMEADPD LSRCPEMVLQ LSLSSATPRS KPVCIHKNSE CLKE QQKRY FPVFSDRVVY TPESLYKELF SQHKGLRDLI NTEMRPFSQG ILIFSQSWAV DLGLQEKQGV ICDALLISQN NTPIL YTIF SKWDAGCKGY SMIVAYSLKQ KLVNKGGYTG RLCITPLVCV LNSDRKAQSV YSSYLQIYPE SYNFMTPQHM EALLQS LVI VLLGFKSFLS EELGSEVLNL LTNKQYELLS KNLRKTRELF VHGLPGSGKT ILALRIMEKI RNVFHCEPAN ILYICEN QP LKKLVSFSKK NICQPVTRKT FMKNNFEHIQ HIIIDDAQNF RTEDGDWYGK AKFITQTARD GPGVLWIFLD YFQTYHLS C SGLPPPSDQY PREEINRVVR NAGPIANYLQ QVMQEARQNP PPNLPPGSLV MLYEPKWAQG VPGNLEIIED LNLEEILIY VANKCRFLLR NGYSPKDIAV LFTKASEVEK YKDRLLTAMR KRKLSQLHEE SDLLLQIGDA SDVLTDHIVL DSVCRFSGLE RNIVFGINP GVAPPAGAYN LLLCLASRAK RHLYILKASV UniProtKB: Schlafen family member 5 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.44 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 140715 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)