+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13175 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Peptide transporter BacA in lipid nanodisc | |||||||||||||||||||||
 Map data Map data | cryoSPARC post-processed map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology | SbmA/BacA-like / SbmA/BacA-like family / : / peptide transport / peptide transmembrane transporter activity / ABC transporter type 1, transmembrane domain superfamily / ATP binding / plasma membrane / Bacteroid development protein BacA Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Sinorhizobium meliloti (bacteria) Sinorhizobium meliloti (bacteria) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.36 Å | |||||||||||||||||||||
 Authors Authors | Ghilarov D / Beis K | |||||||||||||||||||||
| Funding support |  Poland, Poland,  United Kingdom, United Kingdom,  Japan, Japan,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Molecular mechanism of SbmA, a promiscuous transporter exploited by antimicrobial peptides. Authors: Dmitry Ghilarov / Satomi Inaba-Inoue / Piotr Stepien / Feng Qu / Elizabeth Michalczyk / Zuzanna Pakosz / Norimichi Nomura / Satoshi Ogasawara / Graham Charles Walker / Sylvie Rebuffat / So ...Authors: Dmitry Ghilarov / Satomi Inaba-Inoue / Piotr Stepien / Feng Qu / Elizabeth Michalczyk / Zuzanna Pakosz / Norimichi Nomura / Satoshi Ogasawara / Graham Charles Walker / Sylvie Rebuffat / So Iwata / Jonathan Gardiner Heddle / Konstantinos Beis /      Abstract: Antibiotic metabolites and antimicrobial peptides mediate competition between bacterial species. Many of them hijack inner and outer membrane proteins to enter cells. Sensitivity of enteric bacteria ...Antibiotic metabolites and antimicrobial peptides mediate competition between bacterial species. Many of them hijack inner and outer membrane proteins to enter cells. Sensitivity of enteric bacteria to multiple peptide antibiotics is controlled by the single inner membrane protein SbmA. To establish the molecular mechanism of peptide transport by SbmA and related BacA, we determined their cryo–electron microscopy structures at 3.2 and 6 Å local resolution, respectively. The structures show a previously unknown fold, defining a new class of secondary transporters named SbmA-like peptide transporters. The core domain includes conserved glutamates, which provide a pathway for proton translocation, powering transport. The structures show an outward-open conformation with a large cavity that can accommodate diverse substrates. We propose a molecular mechanism for antibacterial peptide uptake paving the way for creation of narrow-targeted therapeutics. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13175.map.gz emd_13175.map.gz | 14.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13175-v30.xml emd-13175-v30.xml emd-13175.xml emd-13175.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
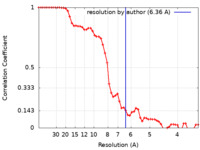
| FSC (resolution estimation) |  emd_13175_fsc.xml emd_13175_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13175.png emd_13175.png | 58.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13175 http://ftp.pdbj.org/pub/emdb/structures/EMD-13175 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13175 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13175 | HTTPS FTP |
-Validation report
| Summary document |  emd_13175_validation.pdf.gz emd_13175_validation.pdf.gz | 352 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13175_full_validation.pdf.gz emd_13175_full_validation.pdf.gz | 351.5 KB | Display | |
| Data in XML |  emd_13175_validation.xml.gz emd_13175_validation.xml.gz | 8.7 KB | Display | |
| Data in CIF |  emd_13175_validation.cif.gz emd_13175_validation.cif.gz | 10.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13175 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13175 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13175 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13175 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13175.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13175.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.69 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Peptide transporter BacA in lipid nanodisc
| Entire | Name: Peptide transporter BacA in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Peptide transporter BacA in lipid nanodisc
| Supramolecule | Name: Peptide transporter BacA in lipid nanodisc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Sinorhizobium meliloti (bacteria) Sinorhizobium meliloti (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Peptide transporter BacA
| Macromolecule | Name: Peptide transporter BacA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sinorhizobium meliloti (bacteria) Sinorhizobium meliloti (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MFQSFFPKPK LFFISSAVWS LLAVLAWYAG GRDIGAYLGL PPLPPGQEPV IGVSVFWSTP FLWFYIYYA VVAGLFAAFW FAYSPHRWQY WSVLGTALII FNTYFSVQVS VAINAWYGPF Y DLIQQALA RTAPVTAGQL YSGMIGFSGI AFVAVTVGVL NLFFVSHYIF ...String: MFQSFFPKPK LFFISSAVWS LLAVLAWYAG GRDIGAYLGL PPLPPGQEPV IGVSVFWSTP FLWFYIYYA VVAGLFAAFW FAYSPHRWQY WSVLGTALII FNTYFSVQVS VAINAWYGPF Y DLIQQALA RTAPVTAGQL YSGMIGFSGI AFVAVTVGVL NLFFVSHYIF RWRTAMNEFY VA HWPRLRH VEGASQRVQE DTMRFSSTVE RLGVGLVSSI MTLIAFLPVL FKFSEQVNVL PIV GEIPHA LVWAAVFWSV FGTVFLAAVG IKLPGLEFRN QRVEAAYRKE LVYGEDHEDR ADPI TLAQL FDNVRRNYFR LYFHYMYFNI ARIFYLQADN LFGTFVLVPA IVAGKLTLGV MNQVL NVFG QVRESFQYLV NSWTTIVELL SIYKRLKAFE SVLVDEPLPE IDRQFIDAGG KEELAL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)