+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13103 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast Sei1 | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Lipid droplet formation / lipid binding / seipin / MEMBRANE PROTEIN | |||||||||||||||||||||||||||
| Function / homology | SEI1 isoform 1 Function and homology information Function and homology information | |||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||||||||
 Authors Authors | Deme JC / Lea SM | |||||||||||||||||||||||||||
| Funding support |  United Kingdom, 8 items United Kingdom, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Authors: Yoel A Klug / Justin C Deme / Robin A Corey / Mike F Renne / Phillip J Stansfeld / Susan M Lea / Pedro Carvalho /   Abstract: Lipid droplets (LDs) are universal lipid storage organelles with a core of neutral lipids, such as triacylglycerols, surrounded by a phospholipid monolayer. This unique architecture is generated ...Lipid droplets (LDs) are universal lipid storage organelles with a core of neutral lipids, such as triacylglycerols, surrounded by a phospholipid monolayer. This unique architecture is generated during LD biogenesis at endoplasmic reticulum (ER) sites marked by Seipin, a conserved membrane protein mutated in lipodystrophy. Here structural, biochemical and molecular dynamics simulation approaches reveal the mechanism of LD formation by the yeast Seipin Sei1 and its membrane partner Ldb16. We show that Sei1 luminal domain assembles a homooligomeric ring, which, in contrast to other Seipins, is unable to concentrate triacylglycerol. Instead, Sei1 positions Ldb16, which concentrates triacylglycerol within the Sei1 ring through critical hydroxyl residues. Triacylglycerol recruitment to the complex is further promoted by Sei1 transmembrane segments, which also control Ldb16 stability. Thus, we propose that LD assembly by the Sei1/Ldb16 complex, and likely other Seipins, requires sequential triacylglycerol-concentrating steps via distinct elements in the ER membrane and lumen. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13103.map.gz emd_13103.map.gz | 179 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13103-v30.xml emd-13103-v30.xml emd-13103.xml emd-13103.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13103_fsc.xml emd_13103_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13103.png emd_13103.png | 184.9 KB | ||
| Masks |  emd_13103_msk_1.map emd_13103_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13103.cif.gz emd-13103.cif.gz | 6.2 KB | ||
| Others |  emd_13103_additional_1.map.gz emd_13103_additional_1.map.gz emd_13103_half_map_1.map.gz emd_13103_half_map_1.map.gz emd_13103_half_map_2.map.gz emd_13103_half_map_2.map.gz | 241.9 MB 243 MB 243.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13103 http://ftp.pdbj.org/pub/emdb/structures/EMD-13103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13103 | HTTPS FTP |
-Related structure data
| Related structure data |  7oxpMC  7oxrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13103.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13103.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13103_msk_1.map emd_13103_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
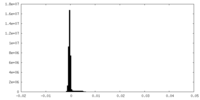
| Density Histograms |
-Additional map: unsharpened map
| File | emd_13103_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
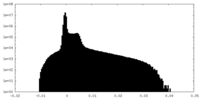
| Density Histograms |
-Half map: unfiltered half map 1
| File | emd_13103_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
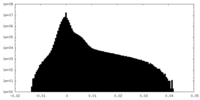
| Density Histograms |
-Half map: unfiltered half map 2
| File | emd_13103_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homodecamer of Sei1
| Entire | Name: homodecamer of Sei1 |
|---|---|
| Components |
|
-Supramolecule #1: homodecamer of Sei1
| Supramolecule | Name: homodecamer of Sei1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: BJ4_G0032880.mRNA.1.CDS.1,BJ4_G0032880.mRNA.1.CDS.1
| Macromolecule | Name: BJ4_G0032880.mRNA.1.CDS.1,BJ4_G0032880.mRNA.1.CDS.1 / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.664125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKINVSRPLQ FLQWSSYIVV AFLIQLLIIL PLSILIYHDF YLRLLPADSS NVVPLNTFNI LNGVQFGTKF FQSIKSIPVG TDLPQTIDN GLSQLIPMRD NMEYKLDLNL QLYCQSKTDH LNLDNLLIDV YRGPGPLLGA PGGSNSKDEK IFHTSRPIVC L ALTDSMSP ...String: MKINVSRPLQ FLQWSSYIVV AFLIQLLIIL PLSILIYHDF YLRLLPADSS NVVPLNTFNI LNGVQFGTKF FQSIKSIPVG TDLPQTIDN GLSQLIPMRD NMEYKLDLNL QLYCQSKTDH LNLDNLLIDV YRGPGPLLGA PGGSNSKDEK IFHTSRPIVC L ALTDSMSP QEIEQLGPSR LDVYDEEWLN TIRIEDKISL ESSYETISVF LKTEIAQRNL IIHPESGIKF RMNFEQGLRN LM LRKRFLS YIIGISIFHC IICVLFFITG CTAFIFVRKG QEKSKKHSGR RIPGLINGGG GGGDYKDHDG DYKDHDIDYK DDD DK UniProtKB: SEI1 isoform 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)