[English] 日本語
 Yorodumi
Yorodumi- EMDB-1290: Lengsin is a survivor of an ancient family of class I glutamine s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1290 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lengsin is a survivor of an ancient family of class I glutamine synthetases re-engineered by evolution for a role in the vertebrate lens. | |||||||||
 Map data Map data | Lengsin | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.0 Å | |||||||||
 Authors Authors | Wyatt K / White HE / Wang L / Bateman OA / Slingsby C / Orlova EV / Wistow G | |||||||||
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Lengsin is a survivor of an ancient family of class I glutamine synthetases re-engineered by evolution for a role in the vertebrate lens. Authors: Keith Wyatt / Helen E White / Luchun Wang / Orval A Bateman / Christine Slingsby / Elena V Orlova / Graeme Wistow /  Abstract: Lengsin is a major protein of the vertebrate eye lens. It belongs to the hitherto purely prokaryotic GS I branch of the glutamine synthetase (GS) superfamily, but has no enzyme activity. Like the ...Lengsin is a major protein of the vertebrate eye lens. It belongs to the hitherto purely prokaryotic GS I branch of the glutamine synthetase (GS) superfamily, but has no enzyme activity. Like the taxon-specific crystallins, Lengsin is the result of the recruitment of an ancient enzyme to a noncatalytic role in the vertebrate lens. Cryo-EM and modeling studies of Lengsin show a dodecamer structure with important similarities and differences with prokaryotic GS I structures. GS homology regions of Lengsin are well conserved, but the N-terminal domain shows evidence of dynamic evolutionary changes. Compared with birds and fish, most mammals have an additional exon corresponding to part of the N-terminal domain; however, in human, this is a nonfunctional pseudoexon. Genes related to Lengsin are also present in the sea urchin, suggesting that this branch of the GS I family, supplanted by GS II enzymes in vertebrates, has an ancient role in metazoans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1290.map.gz emd_1290.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1290-v30.xml emd-1290-v30.xml emd-1290.xml emd-1290.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1290.gif 1290.gif | 18.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1290 http://ftp.pdbj.org/pub/emdb/structures/EMD-1290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1290 | HTTPS FTP |
-Related structure data
| Related structure data |  2j9iMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1290.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1290.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lengsin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
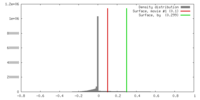
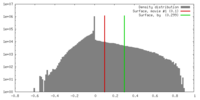
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Lengsin
| Entire | Name: Lengsin |
|---|---|
| Components |
|
-Supramolecule #1000: Lengsin
| Supramolecule | Name: Lengsin / type: sample / ID: 1000 / Oligomeric state: dodecamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 510 KDa / Theoretical: 620.32 KDa |
-Macromolecule #1: Lengsin
| Macromolecule | Name: Lengsin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: dodecamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 6 / Details: 25 mM malonic acid |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Details | Low dose mode |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.32 µm / Nominal defocus min: 0.17 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan single tilt negative stain holder / Specimen holder model: OTHER |
- Image processing
Image processing
| CTF correction | Details: phase flipping |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D6 (2x6 fold dihedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC |
| Final two d classification | Number classes: 150 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: URO |
| Details | Protocol: Rigid Body. The domains were docked manually using COOT |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-2j9i: |
 Movie
Movie Controller
Controller





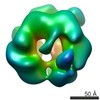



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)