[English] 日本語
 Yorodumi
Yorodumi- EMDB-12766: Single particle cryo-EM reconstruction of just the top two rings ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12766 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM reconstruction of just the top two rings of a 40-mer assembly of recombinant yeast Hsp26 S207E mutant. | |||||||||
 Map data Map data | Cryo-EM Single particle Reconstruction of the top two rings of a 40mer oligomer of yeast Hsp26 S207E mutant, containing 16 monomers. This map was used for flexible fitting of the atomic model | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | small heat shock proteins / Hsp26 S207E mutant / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoplasmic stress granule / unfolded protein binding / cellular response to heat / protein folding / mRNA binding / mitochondrion / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
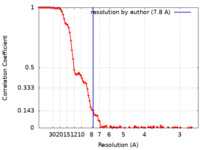
| Method | single particle reconstruction / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | Muehlhofer M / Peters C / Kriehuber T / Kreuzeder M / Kazman P / Rodina N / Reif B / Haslbeck M / Weinkauf S / Buchner J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Phosphorylation activates the yeast small heat shock protein Hsp26 by weakening domain contacts in the oligomer ensemble. Authors: Moritz Mühlhofer / Carsten Peters / Thomas Kriehuber / Marina Kreuzeder / Pamina Kazman / Natalia Rodina / Bernd Reif / Martin Haslbeck / Sevil Weinkauf / Johannes Buchner /  Abstract: Hsp26 is a small heat shock protein (sHsp) from S. cerevisiae. Its chaperone activity is activated by oligomer dissociation at heat shock temperatures. Hsp26 contains 9 phosphorylation sites in ...Hsp26 is a small heat shock protein (sHsp) from S. cerevisiae. Its chaperone activity is activated by oligomer dissociation at heat shock temperatures. Hsp26 contains 9 phosphorylation sites in different structural elements. Our analysis of phospho-mimetic mutations shows that phosphorylation activates Hsp26 at permissive temperatures. The cryo-EM structure of the Hsp26 40mer revealed contacts between the conserved core domain of Hsp26 and the so-called thermosensor domain in the N-terminal part of the protein, which are targeted by phosphorylation. Furthermore, several phosphorylation sites in the C-terminal extension, which link subunits within the oligomer, are sensitive to the introduction of negative charges. In all cases, the intrinsic inhibition of chaperone activity is relieved and the N-terminal domain becomes accessible for substrate protein binding. The weakening of domain interactions within and between subunits by phosphorylation to activate the chaperone activity in response to proteotoxic stresses independent of heat stress could be a general regulation principle of sHsps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12766.map.gz emd_12766.map.gz | 85.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12766-v30.xml emd-12766-v30.xml emd-12766.xml emd-12766.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12766_fsc.xml emd_12766_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12766.png emd_12766.png | 174.4 KB | ||
| Filedesc metadata |  emd-12766.cif.gz emd-12766.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12766 http://ftp.pdbj.org/pub/emdb/structures/EMD-12766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12766 | HTTPS FTP |
-Related structure data
| Related structure data |  7oa6MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12766.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12766.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Single particle Reconstruction of the top two rings of a 40mer oligomer of yeast Hsp26 S207E mutant, containing 16 monomers. This map was used for flexible fitting of the atomic model | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Top two rings (16 subunits) of a 40mer complex of Hsp26 mutant S2...
| Entire | Name: Top two rings (16 subunits) of a 40mer complex of Hsp26 mutant S207E. (use 0.487 as threshold to visualise) |
|---|---|
| Components |
|
-Supramolecule #1: Top two rings (16 subunits) of a 40mer complex of Hsp26 mutant S2...
| Supramolecule | Name: Top two rings (16 subunits) of a 40mer complex of Hsp26 mutant S207E. (use 0.487 as threshold to visualise) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 382 KDa |
-Macromolecule #1: Heat shock protein 26
| Macromolecule | Name: Heat shock protein 26 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 23.910586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFNSPFFDF FDNINNEVDA FNRLLGEGGL RGYAPRRQLA NTPAKDSTGK EVARPNNYAG ALYDPRDETL DDWFDNDLSL FPSGFGFPR SVAVPVDILD HDNNYELKVV VPGVKSKKDI DIEYHQNKNQ ILVSGEIPST LNEESKDKVK VKESSSGKFK R VITLPDYP ...String: MSFNSPFFDF FDNINNEVDA FNRLLGEGGL RGYAPRRQLA NTPAKDSTGK EVARPNNYAG ALYDPRDETL DDWFDNDLSL FPSGFGFPR SVAVPVDILD HDNNYELKVV VPGVKSKKDI DIEYHQNKNQ ILVSGEIPST LNEESKDKVK VKESSSGKFK R VITLPDYP GVDADNIKAD YANGVLTLTV PKLKPQKDGK NHVKKIEVSS QESWGN UniProtKB: Heat shock protein 26 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 5010 / Average exposure time: 8.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Homology model generated with ITASSER with 1GME as template. Fitted with Chimera and flexibly fitted with NAMD. Residues 63 to 214 present in the model. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-7oa6: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)