[English] 日本語
 Yorodumi
Yorodumi- EMDB-12644: Structure of the fungal plasma membrane proton pump Pma1 in its a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12644 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Membrane Protein / P-Type ATPase / proton-transporting ATPase / PROTON TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationP-type H+-exporting transporter / proton export across plasma membrane / P-type proton-exporting transporter activity / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Neurospora crassa (fungus) Neurospora crassa (fungus) | ||||||||||||||||||
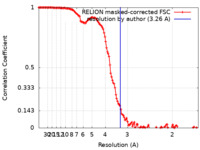
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | ||||||||||||||||||
 Authors Authors | Heit S / Geurts MMG | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state. Authors: Sabine Heit / Maxwell M G Geurts / Bonnie J Murphy / Robin A Corey / Deryck J Mills / Werner Kühlbrandt / Maike Bublitz /   Abstract: The fungal plasma membrane H-ATPase Pma1 is a vital enzyme, generating a proton-motive force that drives the import of essential nutrients. Autoinhibited Pma1 hexamers in the plasma membrane of ...The fungal plasma membrane H-ATPase Pma1 is a vital enzyme, generating a proton-motive force that drives the import of essential nutrients. Autoinhibited Pma1 hexamers in the plasma membrane of starving fungi are activated by glucose signaling and subsequent phosphorylation of the autoinhibitory domain. As related P-type adenosine triphosphatases (ATPases) are not known to oligomerize, the physiological relevance of Pma1 hexamers remained unknown. We have determined the structure of hexameric Pma1 from by electron cryo-microscopy at 3.3-Å resolution, elucidating the molecular basis for hexamer formation and autoinhibition and providing a basis for structure-based drug development. Coarse-grained molecular dynamics simulations in a lipid bilayer suggest lipid-mediated contacts between monomers and a substantial protein-induced membrane deformation that could act as a proton-attracting funnel. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12644.map.gz emd_12644.map.gz | 186.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12644-v30.xml emd-12644-v30.xml emd-12644.xml emd-12644.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12644_fsc.xml emd_12644_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_12644.png emd_12644.png | 192.5 KB | ||
| Masks |  emd_12644_msk_1.map emd_12644_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12644.cif.gz emd-12644.cif.gz | 6.7 KB | ||
| Others |  emd_12644_half_map_1.map.gz emd_12644_half_map_1.map.gz emd_12644_half_map_2.map.gz emd_12644_half_map_2.map.gz | 164.1 MB 164.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12644 http://ftp.pdbj.org/pub/emdb/structures/EMD-12644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12644 | HTTPS FTP |
-Related structure data
| Related structure data |  7ny1MC  7nxfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12644.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12644.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12644_msk_1.map emd_12644_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12644_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12644_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric assembly of the fungal plasma membrane proton pump in i...
| Entire | Name: Hexameric assembly of the fungal plasma membrane proton pump in its auto-inhibited state |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric assembly of the fungal plasma membrane proton pump in i...
| Supramolecule | Name: Hexameric assembly of the fungal plasma membrane proton pump in its auto-inhibited state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) / Strain: FGSC #4761 Neurospora crassa (fungus) / Strain: FGSC #4761 |
-Macromolecule #1: Plasma membrane ATPase
| Macromolecule | Name: Plasma membrane ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: P-type H+-exporting transporter |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 99.984359 KDa |
| Sequence | String: MADHSASGAP ALSTNIESGK FDEKAAEAAA YQPKPKVEDD EDEDIDALIE DLESHDGHDA EEEEEEATPG GGRVVPEDML QTDTRVGLT SEEVVQRRRK YGLNQMKEEK ENHFLKFLGF FVGPIQFVME GAAVLAAGLE DWVDFGVICG LLLLNAVVGF V QEFQAGSI ...String: MADHSASGAP ALSTNIESGK FDEKAAEAAA YQPKPKVEDD EDEDIDALIE DLESHDGHDA EEEEEEATPG GGRVVPEDML QTDTRVGLT SEEVVQRRRK YGLNQMKEEK ENHFLKFLGF FVGPIQFVME GAAVLAAGLE DWVDFGVICG LLLLNAVVGF V QEFQAGSI VDELKKTLAL KAVVLRDGTL KEIEAPEVVP GDILQVEEGT IIPADGRIVT DDAFLQVDQS ALTGESLAVD KH KGDQVFA SSAVKRGEAF VVITATGDNT FVGRAAALVN AASGGSGHFT EVLNGIGTIL LILVIFTLLI VWVSSFYRSN PIV QILEFT LAITIIGVPV GLPAVVTTTM AVGAAYLAKK KAIVQKLSAI ESLAGVEILC SDKTGTLTKN KLSLHDPYTV AGVD PEDLM LTACLAASRK KKGIDAIDKA FLKSLKYYPR AKSVLSKYKV LQFHPFDPVS KKVVAVVESP QGERITCVKG APLFV LKTV EEDHPIPEEV DQAYKNKVAE FATRGFRSLG VARKRGEGSW EILGIMPCMD PPRHDTYKTV CEAKTLGLSI KMLTGD AVG IARETSRQLG LGTNIYNAER LGLGGGGDMP GSEVYDFVEA ADGFAEVFPQ HKYNVVEILQ QRGYLVAMTG DGVNDAP SL KKADTGIAVE GSSDAARSAA DIVFLAPGLG AIIDALKTSR QIFHRMYAYV VYRIALSIHL EIFLGLWIAI LNRSLNIE L VVFIAIFADV ATLAIAYDNA PYSQTPVKWN LPKLWGMSVL LGVVLAVGTW ITVTTMYAQG ENGGIVQNFG NMDEVLFLQ ISLTENWLIF ITRANGPFWS SIPSWQLSGA IFLVDILATC FTIWGWFEHS DTSIVAVVRI WIFSFGIFCI MGGVYYILQD SVGFDNLMH GKSPKGNQKQ RSLEDFVVSL QRVSTQHEKS Q UniProtKB: Plasma membrane ATPase |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)