+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12602 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Pol Kappa holoenzyme with Ub-PCNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Translesion synthesis / TLS / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication / purine-specific mismatch base pair DNA N-glycosylase activity / nucleotide-excision repair, DNA gap filling / nuclear lamina / Polymerase switching / positive regulation of DNA-directed DNA polymerase activity / Processive synthesis on the lagging strand ...positive regulation of deoxyribonuclease activity / dinucleotide insertion or deletion binding / PCNA-p21 complex / mitotic telomere maintenance via semi-conservative replication / purine-specific mismatch base pair DNA N-glycosylase activity / nucleotide-excision repair, DNA gap filling / nuclear lamina / Polymerase switching / positive regulation of DNA-directed DNA polymerase activity / Processive synthesis on the lagging strand / MutLalpha complex binding / PCNA complex / Telomere C-strand (Lagging Strand) Synthesis / Removal of the Flap Intermediate / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Transcription of E2F targets under negative control by DREAM complex / Polymerase switching on the C-strand of the telomere / replisome / Processive synthesis on the C-strand of the telomere / response to L-glutamate / Removal of the Flap Intermediate from the C-strand / response to dexamethasone / histone acetyltransferase binding / DNA polymerase processivity factor activity / leading strand elongation / G1/S-Specific Transcription / nuclear replication fork / replication fork processing / SUMOylation of DNA replication proteins / PCNA-Dependent Long Patch Base Excision Repair / error-prone translesion synthesis / response to cadmium ion / estrous cycle / mismatch repair / translesion synthesis / cyclin-dependent protein kinase holoenzyme complex / base-excision repair, gap-filling / DNA polymerase binding / epithelial cell differentiation / liver regeneration / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / positive regulation of DNA replication / nuclear estrogen receptor binding / replication fork / Translesion synthesis by REV1 / positive regulation of DNA repair / Translesion synthesis by POLK / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / male germ cell nucleus / Termination of translesion DNA synthesis / Translesion Synthesis by POLH / Recognition of DNA damage by PCNA-containing replication complex / receptor tyrosine kinase binding / HDR through Homologous Recombination (HRR) / cellular response to xenobiotic stimulus / Dual Incision in GG-NER / cellular response to hydrogen peroxide / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / cellular response to UV / response to estradiol / E3 ubiquitin ligases ubiquitinate target proteins / heart development / chromatin organization / DNA-directed DNA polymerase / damaged DNA binding / DNA-directed DNA polymerase activity / DNA replication / chromosome, telomeric region / nuclear body / DNA repair / DNA damage response / chromatin binding / centrosome / chromatin / protein-containing complex binding / enzyme binding / negative regulation of transcription by RNA polymerase II / extracellular exosome / zinc ion binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
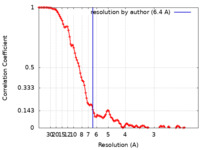
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||
 Authors Authors | Lancey C / De Biasio A | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structure of human Pol κ bound to DNA and mono-ubiquitylated PCNA. Authors: Claudia Lancey / Muhammad Tehseen / Souvika Bakshi / Matthew Percival / Masateru Takahashi / Mohamed A Sobhy / Vlad S Raducanu / Kerry Blair / Frederick W Muskett / Timothy J Ragan / Ramon ...Authors: Claudia Lancey / Muhammad Tehseen / Souvika Bakshi / Matthew Percival / Masateru Takahashi / Mohamed A Sobhy / Vlad S Raducanu / Kerry Blair / Frederick W Muskett / Timothy J Ragan / Ramon Crehuet / Samir M Hamdan / Alfredo De Biasio /    Abstract: Y-family DNA polymerase κ (Pol κ) can replicate damaged DNA templates to rescue stalled replication forks. Access of Pol κ to DNA damage sites is facilitated by its interaction with the ...Y-family DNA polymerase κ (Pol κ) can replicate damaged DNA templates to rescue stalled replication forks. Access of Pol κ to DNA damage sites is facilitated by its interaction with the processivity clamp PCNA and is regulated by PCNA mono-ubiquitylation. Here, we present cryo-EM reconstructions of human Pol κ bound to DNA, an incoming nucleotide, and wild type or mono-ubiquitylated PCNA (Ub-PCNA). In both reconstructions, the internal PIP-box adjacent to the Pol κ Polymerase-Associated Domain (PAD) docks the catalytic core to one PCNA protomer in an angled orientation, bending the DNA exiting the Pol κ active site through PCNA, while Pol κ C-terminal domain containing two Ubiquitin Binding Zinc Fingers (UBZs) is invisible, in agreement with disorder predictions. The ubiquitin moieties are partly flexible and extend radially away from PCNA, with the ubiquitin at the Pol κ-bound protomer appearing more rigid. Activity assays suggest that, when the internal PIP-box interaction is lost, Pol κ is retained on DNA by a secondary interaction between the UBZs and the ubiquitins flexibly conjugated to PCNA. Our data provide a structural basis for the recruitment of a Y-family TLS polymerase to sites of DNA damage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12602.map.gz emd_12602.map.gz | 8.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12602-v30.xml emd-12602-v30.xml emd-12602.xml emd-12602.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12602_fsc.xml emd_12602_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
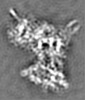
| Images |  emd_12602.png emd_12602.png | 62.1 KB | ||
| Filedesc metadata |  emd-12602.cif.gz emd-12602.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12602 http://ftp.pdbj.org/pub/emdb/structures/EMD-12602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12602 | HTTPS FTP |
-Related structure data
| Related structure data |  7nv1MC  7nv0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10934 (Title: Cryo-EM multiframe micrographs of human Pol kappa complexed with DNA and monoubiquitylated PCNA EMPIAR-10934 (Title: Cryo-EM multiframe micrographs of human Pol kappa complexed with DNA and monoubiquitylated PCNAData size: 1.9 TB Data #1: Multiframe micrographs of human Pol κ complexed with monoubiquitylated PCNA and DNA [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12602.map.gz / Format: CCP4 / Size: 9.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12602.map.gz / Format: CCP4 / Size: 9.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.086 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pol kappa holoenzyme with Ub-PCNA
| Entire | Name: Pol kappa holoenzyme with Ub-PCNA |
|---|---|
| Components |
|
-Supramolecule #1: Pol kappa holoenzyme with Ub-PCNA
| Supramolecule | Name: Pol kappa holoenzyme with Ub-PCNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 232 KDa |
-Supramolecule #2: Pol kappa holoenzyme
| Supramolecule | Name: Pol kappa holoenzyme / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Proliferating cell nuclear antigen
| Macromolecule | Name: Proliferating cell nuclear antigen / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.088061 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPHMFEARLV QGSILKKVLE ALKDLINEAC WDISSSGVNL QSMDSSHVSL VQLTLRSEGF DTYRCDRNLA MGVNLTSMSK ILKCAGNED IITLRAEDNA DTLALVFEAP NQEKVSDYEM KLMDLDVEQL GIPEQEYSCV VKMPSGEFAR ICRDLSHIGD A VVISCAKD ...String: GPHMFEARLV QGSILKKVLE ALKDLINEAC WDISSSGVNL QSMDSSHVSL VQLTLRSEGF DTYRCDRNLA MGVNLTSMSK ILKCAGNED IITLRAEDNA DTLALVFEAP NQEKVSDYEM KLMDLDVEQL GIPEQEYSCV VKMPSGEFAR ICRDLSHIGD A VVISCAKD GVKFSASGEL GNGNIKLSQT SNVDKEEEAV TIEMNEPVQL TFALRYLNFF TKATPLSSTV TLSMSADVPL VV EYKIADM GHLKYYLAPK IEDEEGS UniProtKB: Proliferating cell nuclear antigen |
-Macromolecule #2: DNA polymerase kappa
| Macromolecule | Name: DNA polymerase kappa / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.952695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSTKEKCDS YKDDLLLRMG LNDNKAGMEG LDKEKINKII MEATKGSRFY GNELKKEKQV NQRIENMMQQ KAQITSQQLR KAQLQVDRF AMELEQSRNL SNTIVHIDMD AFYAAVEMRD NPELKDKPIA VGSMSMLSTS NYHARRFGVR AAMPGFIAKR L CPQLIIVP ...String: MDSTKEKCDS YKDDLLLRMG LNDNKAGMEG LDKEKINKII MEATKGSRFY GNELKKEKQV NQRIENMMQQ KAQITSQQLR KAQLQVDRF AMELEQSRNL SNTIVHIDMD AFYAAVEMRD NPELKDKPIA VGSMSMLSTS NYHARRFGVR AAMPGFIAKR L CPQLIIVP PNFDKYRAVS KEVKEILADY DPNFMAMSLD EAYLNITKHL EERQNWPEDK RRYFIKMGSS VENDNPGKEV NK LSEHERS ISPLLFEESP SDVQPPGDPF QVNFEEQNNP QILQNSVVFG TSAQEVVKEI RFRIEQKTTL TASAGIAPNT MLA KVCSDK NKPNGQYQIL PNRQAVMDFI KDLPIRKVSG IGKVTEKMLK ALGIITCTEL YQQRALLSLL FSETSWHYFL HISL GLGST HLTRDGERKS MSVERTFSEI NKAEEQYSLC QELCSELAQD LQKERLKGRT VTIKLKNVNF EVKTRASTVS SVVST AEEI FAIAKELLKT EIDADFPHPL RLRLMGVRIS SFPNEEDRKH QQRSIIGFLQ AGNQALSATE CTLEKTDKDK FVKPLE MSH KKSFFDKKRS ERKWSHQDTF KCEAVNKQSF QTSQPFQVLK KKMNENLEIS ENSDDCQILT CPVCFRAQGC ISLEALN KH VDECLDGPSI SENFKMFSCS HVSATKVNKK ENVPASSLCE KQDYEAHPKI KEISSVDCIA LVDTIDNSSK AESIDALS N KHSKEECSSL PSKSFNIEHC HQNSSSTVSL ENEDVGSFRQ EYRQPYLCEV KTGQALVCPV CNVEQKTSDL TLFNVHVDV CLNKSFIQEL RKDKFNPVNQ PKESSRSTGS SSGVQKAVTR TKRPGLMTKY STSKKIKPNN PKHTLDIFFK UniProtKB: DNA polymerase kappa |
-Macromolecule #3: DNA Primer
| Macromolecule | Name: DNA Primer / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.665987 KDa |
| Sequence | String: (DA)(DG)(DC)(DT)(DA)(DT)(DG)(DA)(DC)(DC) (DA)(DT)(DG)(DA)(DT)(DT)(DA)(DC)(DG)(DA) (DA)(DT)(DT)(DG)(DOC) |
-Macromolecule #4: DNA Template
| Macromolecule | Name: DNA Template / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 11.677539 KDa |
| Sequence | String: (DC)(DT)(DG)(DC)(DA)(DC)(DG)(DA)(DA)(DT) (DT)(DA)(DA)(DG)(DC)(DA)(DA)(DT)(DT)(DC) (DG)(DT)(DA)(DA)(DT)(DC)(DA)(DT)(DG) (DG)(DT)(DC)(DA)(DT)(DA)(DG)(DC)(DT) |
-Macromolecule #5: THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: TTP |
|---|---|
| Molecular weight | Theoretical: 482.168 Da |
| Chemical component information |  ChemComp-TTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. / Details: 40 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 3889 / Average exposure time: 5.0 sec. / Average electron dose: 47.0 e/Å2 / Details: Super resolution mode & AFIS used |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7nv1: |
 Movie
Movie Controller
Controller































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)