[English] 日本語
 Yorodumi
Yorodumi- EMDB-1212: Hrr25-dependent phosphorylation state regulates organization of t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1212 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. | |||||||||
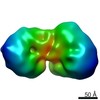
 Map data Map data | map of 40S ribosome | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Schafer T / Maco B / Petfalski E / Tollervey D / Bottcher B / Aebi U / Hurt E | |||||||||
 Citation Citation |  Journal: Nature / Year: 2006 Journal: Nature / Year: 2006Title: Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Authors: Thorsten Schäfer / Bohumil Maco / Elisabeth Petfalski / David Tollervey / Bettina Böttcher / Ueli Aebi / Ed Hurt /  Abstract: The formation of eukaryotic ribosomes is a multistep process that takes place successively in the nucleolar, nucleoplasmic and cytoplasmic compartments. Along this pathway, multiple pre-ribosomal ...The formation of eukaryotic ribosomes is a multistep process that takes place successively in the nucleolar, nucleoplasmic and cytoplasmic compartments. Along this pathway, multiple pre-ribosomal particles are generated, which transiently associate with numerous non-ribosomal factors before mature 60S and 40S subunits are formed. However, most mechanistic details of ribosome biogenesis are still unknown. Here we identify a maturation step of the yeast pre-40S subunit that is regulated by the protein kinase Hrr25 and involves ribosomal protein Rps3. A high salt concentration releases Rps3 from isolated pre-40S particles but not from mature 40S subunits. Electron microscopy indicates that pre-40S particles lack a structural landmark present in mature 40S subunits, the 'beak'. The beak is formed by the protrusion of 18S ribosomal RNA helix 33, which is in close vicinity to Rps3. Two protein kinases Hrr25 and Rio2 are associated with pre-40S particles. Hrr25 phosphorylates Rps3 and the 40S synthesis factor Enp1. Phosphorylated Rsp3 and Enp1 readily dissociate from the pre-ribosome, whereas subsequent dephosphorylation induces formation of the beak structure and salt-resistant integration of Rps3 into the 40S subunit. In vivo depletion of Hrr25 inhibits growth and leads to the accumulation of immature 40S subunits that contain unstably bound Rps3. We conclude that the kinase activity of Hrr25 regulates the maturation of 40S ribosomal subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1212.map.gz emd_1212.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1212-v30.xml emd-1212-v30.xml emd-1212.xml emd-1212.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  1212.gif 1212.gif | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1212 http://ftp.pdbj.org/pub/emdb/structures/EMD-1212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1212 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1212.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1212.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of 40S ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 40S Ribosome
| Entire | Name: 40S Ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: 40S Ribosome
| Supramolecule | Name: 40S Ribosome / type: sample / ID: 1000 / Details: sample was purified from S. cervisiae / Oligomeric state: monomer / Number unique components: 1 |
|---|
-Supramolecule #1: 40S-ribosome
| Supramolecule | Name: 40S-ribosome / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-eukaryote: SSU 40S |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50mM Tris-HCl, 100mM NaCl, 10mM MgCl2, 0.5mM DTT |
|---|---|
| Grid | Details: 400 mesh copper rhodium grids (Maxtaform) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Plunger with environmental chamber Method: Blot for 15 s with Whatman No 1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 94 K |
| Alignment procedure | Legacy - Astigmatism: bjective lens astigmatism was corrected at |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 14 µm / Number real images: 69 / Bits/pixel: 12 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.2 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















