[English] 日本語
 Yorodumi
Yorodumi- EMDB-12089: Acinetobacter baumannii multidrug transporter AdeB in L*OO state -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12089 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acinetobacter baumannii multidrug transporter AdeB in L*OO state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | RND-transporter / multidrug transporter / antibiotic resistance / membrane protein / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology | Hydrophobe/amphiphile efflux-1 HAE1 / Multidrug efflux transporter AcrB TolC docking domain, DN/DC subdomains / Acriflavin resistance protein / AcrB/AcrD/AcrF family / efflux transmembrane transporter activity / xenobiotic transport / response to toxic substance / plasma membrane / Efflux pump membrane transporter Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Acinetobacter baumannii AYE (bacteria) / Acinetobacter baumannii AYE (bacteria) /  Acinetobacter baumannii (strain AYE) (bacteria) Acinetobacter baumannii (strain AYE) (bacteria) | ||||||||||||||||||
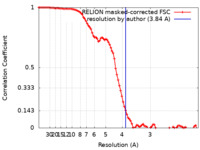
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | ||||||||||||||||||
 Authors Authors | Ornik-Cha A / Reitz J | ||||||||||||||||||
| Funding support |  Germany, European Union, 5 items Germany, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms. Authors: Alina Ornik-Cha / Julia Wilhelm / Jessica Kobylka / Hanno Sjuts / Attilio V Vargiu / Giuliano Malloci / Julian Reitz / Anja Seybert / Achilleas S Frangakis / Klaas M Pos /   Abstract: Upon antibiotic stress Gram-negative pathogens deploy resistance-nodulation-cell division-type tripartite efflux pumps. These include a H/drug antiporter module that recognizes structurally diverse ...Upon antibiotic stress Gram-negative pathogens deploy resistance-nodulation-cell division-type tripartite efflux pumps. These include a H/drug antiporter module that recognizes structurally diverse substances, including antibiotics. Here, we show the 3.5 Å structure of subunit AdeB from the Acinetobacter baumannii AdeABC efflux pump solved by single-particle cryo-electron microscopy. The AdeB trimer adopts mainly a resting state with all protomers in a conformation devoid of transport channels or antibiotic binding sites. However, 10% of the protomers adopt a state where three transport channels lead to the closed substrate (deep) binding pocket. A comparison between drug binding of AdeB and Escherichia coli AcrB is made via activity analysis of 20 AdeB variants, selected on basis of side chain interactions with antibiotics observed in the AcrB periplasmic domain X-ray co-structures with fusidic acid (2.3 Å), doxycycline (2.1 Å) and levofloxacin (2.7 Å). AdeABC, compared to AcrAB-TolC, confers higher resistance to E. coli towards polyaromatic compounds and lower resistance towards antibiotic compounds. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12089.map.gz emd_12089.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12089-v30.xml emd-12089-v30.xml emd-12089.xml emd-12089.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12089_fsc.xml emd_12089_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12089.png emd_12089.png | 158 KB | ||
| Filedesc metadata |  emd-12089.cif.gz emd-12089.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12089 http://ftp.pdbj.org/pub/emdb/structures/EMD-12089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12089 | HTTPS FTP |
-Related structure data
| Related structure data |  7b8qMC  7b8pC  7b8rC  7b8sC  7b8tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12089.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12089.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Homotrimeric RND-transporter AdeB reconstituted in Salipro
| Entire | Name: Homotrimeric RND-transporter AdeB reconstituted in Salipro |
|---|---|
| Components |
|
-Supramolecule #1: Homotrimeric RND-transporter AdeB reconstituted in Salipro
| Supramolecule | Name: Homotrimeric RND-transporter AdeB reconstituted in Salipro type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AYE (bacteria) Acinetobacter baumannii AYE (bacteria) |
| Molecular weight | Theoretical: 345 KDa |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (strain AYE) (bacteria) / Strain: AYE Acinetobacter baumannii (strain AYE) (bacteria) / Strain: AYE |
| Molecular weight | Theoretical: 115.143055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSMSQFFIRR PVFAWVIAIF IIIFGLLSIP KLPIARFPSV APPQVNISAT YPGATAKTIN DSVVTLIERE LSGVKNLLYY SATTDTSGT AEITATFKPG TDVEMAQVDV QNKIKAVEAR LPQVVRQQGL QVEASSSGFL MLVGINSPNN QYSEVDLSDY L VRNVVEEL ...String: MSMSQFFIRR PVFAWVIAIF IIIFGLLSIP KLPIARFPSV APPQVNISAT YPGATAKTIN DSVVTLIERE LSGVKNLLYY SATTDTSGT AEITATFKPG TDVEMAQVDV QNKIKAVEAR LPQVVRQQGL QVEASSSGFL MLVGINSPNN QYSEVDLSDY L VRNVVEEL KRVEGVGKVQ SFGAEKAMRI WVDPNKLVSY GLSISDVNNA IRENNVEIAP GRLGDLPAEK GQLITIPLSA QG QLSSLEQ FKNISLKSKT NGSVIKLSDV ANVEIGSQAY NFAILENGKP ATAAAIQLSP GANAVKTAEG VRAKIEELKL NLP EGMEFS IPYDTAPFVK ISIEKVIHTL LEAMVLVFIV MYLFLHNVRY TLIPAIVAPI ALLGTFTVML LAGFSINVLT MFGM VLAIG IIVDDAIVVV ENVERIMATE GLSPKDATSK AMKEITSPII GITLVLAAVF LPMAFASGSV GVIYKQFTLT MSVSI LFSA LLALILTPAL CATILKPIDG HHQKKGFFAW FDRSFDKVTK KYELMLLKII KHTVPMMVIF LVITGITFAG MKYWPT AFM PEEDQGWFMT SFQLPSDATA ERTRNVVNQF ENNLKDNPDV KSNTAILGWG FSGAGQNVAV AFTTLKDFKE RTSSASK MT SDVNSSMANS TEGETMAVLP PAIDELGTFS GFSLRLQDRA NLGMPALLAA QDELMAMAAK NKKFYMVWNE GLPQGDNI S LKIDREKLSA LGVKFSDVSD IISTSMGSMY INDFPNQGRM QQVIVQVEAK SRMQLKDILN LKVMGSSGQL VSLSEVVTP QWNKAPQQYN RYNGRPSLSI AGIPNFDTSS GEAMREMEQL IAKLPKGIGY EWTGISLQEK QSESQMAFLL GLSMLVVFLV LAALYESWA IPLSVMLVVP LGIFGAIIAI MSRGLMNDVF FKIGLITIIG LSAKNAILIV EFAKMLKEEG MSLIEATVAA A KLRLRPIL MTSLAFTCGV IPLVIATGAS SETQHALGTG VFGGMISATI LAIFFVPVFF IFILGAVEKL FSSKKKISSA LE VLFQGPH HHHHHHHHH UniProtKB: Efflux pump membrane transporter |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7b8q: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)