[English] 日本語
 Yorodumi
Yorodumi- EMDB-1208: Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1208 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
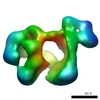
| Title | Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. | |||||||||
 Map data Map data | 3D structure of the DNA-bound DNAPKcs/Ku70/ku80 complex using cryoEM | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | : / Ku70 / Phosphatidylinositol 3-/4-kinase, catalytic domain / double-strand break repair via nonhomologous end joining / double-strand break repair / protein kinase activity Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Spagnolo L / Rivera-Calzada A / Pearl LH / Llorca O | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2006 Journal: Mol Cell / Year: 2006Title: Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Authors: Laura Spagnolo / Angel Rivera-Calzada / Laurence H Pearl / Oscar Llorca /  Abstract: DNA-PKcs is a large (approximately 470 kDa) kinase that plays an essential role in the repair of DNA double-strand breaks (DSBs) by nonhomologous end joining (NHEJ). DNA-PKcs is recruited to DSBs by ...DNA-PKcs is a large (approximately 470 kDa) kinase that plays an essential role in the repair of DNA double-strand breaks (DSBs) by nonhomologous end joining (NHEJ). DNA-PKcs is recruited to DSBs by the Ku70/Ku80 heterodimer, with which it forms the core of a multiprotein complex that promotes synapsis of the broken DNA ends. We have purified the human DNA-PKcs/Ku70/Ku80 holoenzyme assembled on a DNA molecule. Its three-dimensional (3D) structure at approximately 25 Angstroms resolution was determined by single-particle electron microscopy. Binding of Ku and DNA elicits conformational changes in the FAT and FATC domains of DNA-PKcs. Dimeric particles are observed in which two DNA-PKcs/Ku70/Ku80 holoenzymes interact through the N-terminal HEAT repeats. The proximity of the dimer contacts to the likely positions of the DNA ends suggests that these represent synaptic complexes that maintain broken DNA ends in proximity and provide a platform for access of the various enzymes required for end processing and ligation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1208.map.gz emd_1208.map.gz | 990.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1208-v30.xml emd-1208-v30.xml emd-1208.xml emd-1208.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1208.gif 1208.gif | 61.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1208 http://ftp.pdbj.org/pub/emdb/structures/EMD-1208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1208 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1208.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1208.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D structure of the DNA-bound DNAPKcs/Ku70/ku80 complex using cryoEM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Macromolecular Complex containing DNA, DNAPKcs, Ku70 and Ku80
| Entire | Name: Macromolecular Complex containing DNA, DNAPKcs, Ku70 and Ku80 |
|---|---|
| Components |
|
-Supramolecule #1000: Macromolecular Complex containing DNA, DNAPKcs, Ku70 and Ku80
| Supramolecule | Name: Macromolecular Complex containing DNA, DNAPKcs, Ku70 and Ku80 type: sample / ID: 1000 Oligomeric state: One molecule of DNAPKcs and one Ku70-Ku80 dimer bound to one DNA molecule Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: DNA-PKcs
| Macromolecule | Name: DNA-PKcs / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus / Location in cell: Nucleus |
| Molecular weight | Experimental: 470 KDa |
| Recombinant expression | Organism: HeLa cells |
| Sequence | GO: protein kinase activity InterPro: Phosphatidylinositol 3-/4-kinase, catalytic domain |
-Macromolecule #2: Ku70
| Macromolecule | Name: Ku70 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus Homo sapiens (human) / synonym: Human / Cell: HeLa / Organelle: Nucleus |
| Molecular weight | Experimental: 70 KDa |
| Recombinant expression | Organism: HeLa cells |
| Sequence | GO: double-strand break repair via nonhomologous end joining InterPro: Ku70 |
-Macromolecule #3: Ku80
| Macromolecule | Name: Ku80 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: HeLa cells / Organelle: Nucleus Homo sapiens (human) / synonym: Human / Cell: HeLa cells / Organelle: Nucleus |
| Molecular weight | Experimental: 85 KDa |
| Recombinant expression | Organism: HeLa cells |
| Sequence | GO: double-strand break repair / InterPro: INTERPRO: IPR011210 |
-Macromolecule #4: DNA
| Macromolecule | Name: DNA / type: dna / ID: 4 / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 20 mM Hepes, 200 mM KCl, 0.1 mM DTT, 0.5 mM EDTA |
|---|---|
| Grid | Details: 400 mesh carbon coated Rhodium-Copper grids |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home made plunger / Method: Blot for a few seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: FFT with ssCCD |
| Details | Microscope model: TECNAI G2 200 kV, FEI. |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 10 µm / Number real images: 32 / Details: Scanner: MINOLTA Dimage Scan Multi Pro / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: side entry liquid-nitrogen cooled cryo specimen holder. Eucentric Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each partcile using CTFIND3 and EMAN. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 4189 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















