+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3607 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SWR1 chromatin remodelling complex | |||||||||
 Map data Map data | SWR1 chromatin remodelling complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 21.0 Å | |||||||||
 Authors Authors | Chaban Y / Lin C-L / Wigley DB | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2017 Journal: Nucleic Acids Res / Year: 2017Title: Functional characterization and architecture of recombinant yeast SWR1 histone exchange complex. Authors: Chia-Liang Lin / Yuriy Chaban / David M Rees / Elizabeth A McCormack / Lorraine Ocloo / Dale B Wigley /  Abstract: We have prepared recombinant fourteen subunit yeast SWR1 complex from insect cells using a modified MultiBac system. The 1.07 MDa recombinant protein complex has histone-exchange activity. Full ...We have prepared recombinant fourteen subunit yeast SWR1 complex from insect cells using a modified MultiBac system. The 1.07 MDa recombinant protein complex has histone-exchange activity. Full exchange activity is realized with a single SWR1 complex bound to a nucleosome. We also prepared mutant complexes that lack a variety of subunits or combinations of subunits and these start to reveal roles for some of these subunits as well as indicating interactions between them in the full complex. Complexes containing a series of N-terminally and C-terminally truncated Swr1 subunits reveal further details about interactions between subunits as well as their binding sites on the Swr1 subunit. Finally, we present electron microscopy studies revealing the dynamic nature of the complex and a 21 Å resolution reconstruction of the intact complex provides details not apparent in previously reported structures, including a large central cavity of sufficient size to accommodate a nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3607.map.gz emd_3607.map.gz | 1008.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3607-v30.xml emd-3607-v30.xml emd-3607.xml emd-3607.xml | 7.8 KB 7.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3607.png emd_3607.png | 80.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3607 http://ftp.pdbj.org/pub/emdb/structures/EMD-3607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3607 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3607 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3607.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3607.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SWR1 chromatin remodelling complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
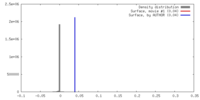
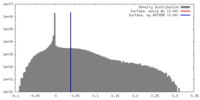
| Voxel size | X=Y=Z: 2.75 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SWR1 chromatin remodelling complex
| Entire | Name: SWR1 chromatin remodelling complex |
|---|---|
| Components |
|
-Supramolecule #1: SWR1 chromatin remodelling complex
| Supramolecule | Name: SWR1 chromatin remodelling complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER Details: Initial 3D model was obtained from 2D class averages using angular reconstitution technique. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 21.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 6000 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)