[English] 日本語
 Yorodumi
Yorodumi- EMDB-11991: Cryo-EM density map of complement C4b in complex with nanobody G3 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11991 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM density map of complement C4b in complex with nanobody G3 | |||||||||
 Map data Map data | DeepEMhancer-postprocessed, local-sharpened cryo-EM density map of complement C4b in complex with nanobody G3. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcomplement activation, GZMK pathway / detection of molecule of bacterial origin / complement binding / opsonization / symbiont cell surface / positive regulation of apoptotic cell clearance / Activation of C3 and C5 / complement activation / endopeptidase inhibitor activity / Initial triggering of complement ...complement activation, GZMK pathway / detection of molecule of bacterial origin / complement binding / opsonization / symbiont cell surface / positive regulation of apoptotic cell clearance / Activation of C3 and C5 / complement activation / endopeptidase inhibitor activity / Initial triggering of complement / complement activation, classical pathway / Regulation of Complement cascade / carbohydrate binding / blood microparticle / inflammatory response / axon / synapse / dendrite / cell surface / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
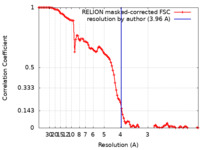
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Oosterheert W / De la O Becerra KI / Gros P | |||||||||
| Funding support |  Mexico, 1 items Mexico, 1 items
| |||||||||
 Citation Citation |  Journal: J Immunol / Year: 2022 Journal: J Immunol / Year: 2022Title: Multifaceted Activities of Seven Nanobodies against Complement C4b. Authors: Karla I De la O Becerra / Wout Oosterheert / Ramon M van den Bos / Katerina T Xenaki / Joseph H Lorent / Maartje Ruyken / Arie Schouten / Suzan H M Rooijakkers / Paul M P van Bergen En Henegouwen / Piet Gros /  Abstract: Cleavage of the mammalian plasma protein C4 into C4b initiates opsonization, lysis, and clearance of microbes and damaged host cells by the classical and lectin pathways of the complement system. ...Cleavage of the mammalian plasma protein C4 into C4b initiates opsonization, lysis, and clearance of microbes and damaged host cells by the classical and lectin pathways of the complement system. Dysregulated activation of C4 and other initial components of the classical pathway may cause or aggravate pathologies, such as systemic lupus erythematosus, Alzheimer disease, and schizophrenia. Modulating the activity of C4b by small-molecule or protein-based inhibitors may represent a promising therapeutic approach for preventing excessive inflammation and damage to host cells and tissue. Here, we present seven nanobodies, derived from llama () immunization, that bind to human C4b () with high affinities ranging from 3.2 nM to 14 pM. The activity of the nanobodies varies from no to complete inhibition of the classical pathway. The inhibiting nanobodies affect different steps in complement activation, in line with blocking sites for proconvertase formation, C3 substrate binding to the convertase, and regulator-mediated inactivation of C4b. For four nanobodies, we determined single-particle cryo-electron microscopy structures in complex with C4b at 3.4-4 Å resolution. The structures rationalize the observed functional effects of the nanobodies and define their mode of action during complement activation. Thus, we characterized seven anti-C4b nanobodies with diverse effects on the classical pathway of complement activation that may be explored for imaging, diagnostic, or therapeutic applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11991.map.gz emd_11991.map.gz | 91.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11991-v30.xml emd-11991-v30.xml emd-11991.xml emd-11991.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11991_fsc.xml emd_11991_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11991.png emd_11991.png | 88.9 KB | ||
| Masks |  emd_11991_msk_1.map emd_11991_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_11991_additional_1.map.gz emd_11991_additional_1.map.gz emd_11991_additional_2.map.gz emd_11991_additional_2.map.gz emd_11991_half_map_1.map.gz emd_11991_half_map_1.map.gz emd_11991_half_map_2.map.gz emd_11991_half_map_2.map.gz | 80.6 MB 62.9 MB 80.8 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11991 http://ftp.pdbj.org/pub/emdb/structures/EMD-11991 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11991 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11991 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11991.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11991.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer-postprocessed, local-sharpened cryo-EM density map of complement C4b in complex with nanobody G3. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0285 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11991_msk_1.map emd_11991_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Refined, unsharpened cryo-EM density map of complement C4b...
| File | emd_11991_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined, unsharpened cryo-EM density map of complement C4b in complex with nanobody G3. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local-resolution filtered, sharpened cryo-EM density map of complement...
| File | emd_11991_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-resolution filtered, sharpened cryo-EM density map of complement C4b in complex with nanobody G3. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of the refinement of the...
| File | emd_11991_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of the refinement of the cryo-EM density map of complement C4b in complex with nanobody G3. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of the refinement of the...
| File | emd_11991_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of the refinement of the cryo-EM density map of complement C4b in complex with nanobody G3. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complement C4b bound to nanobody G3
| Entire | Name: Complement C4b bound to nanobody G3 |
|---|---|
| Components |
|
-Supramolecule #1: Complement C4b bound to nanobody G3
| Supramolecule | Name: Complement C4b bound to nanobody G3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 14.4 KDa |
-Supramolecule #2: Complement C4b
| Supramolecule | Name: Complement C4b / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: The purchased C4b is generated from a mixture of C4A and C4B. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Nanobody G3
| Supramolecule | Name: Nanobody G3 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Complement C4b
| Macromolecule | Name: Complement C4b / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRLLWGLIWA SSFFTLSLQK PRLLLFSPSV VHLGVPLSVG VQLQDVPRGQ VVKGSVFLRN PSRNNVPCS PKVDFTLSSE RDFALLSLQV PLKDAKSCGL HQLLRGPEVQ LVAHSPWLKD S LSRTTNIQ GINLLFSSRR GHLFLQTDQP IYNPGQRVRY RVFALDQKMR ...String: MRLLWGLIWA SSFFTLSLQK PRLLLFSPSV VHLGVPLSVG VQLQDVPRGQ VVKGSVFLRN PSRNNVPCS PKVDFTLSSE RDFALLSLQV PLKDAKSCGL HQLLRGPEVQ LVAHSPWLKD S LSRTTNIQ GINLLFSSRR GHLFLQTDQP IYNPGQRVRY RVFALDQKMR PSTDTITVMV EN SHGLRVR KKEVYMPSSI FQDDFVIPDI SEPGTWKISA RFSDGLESNS STQFEVKKYV LPN FEVKIT PGKPYILTVP GHLDEMQLDI QARYIYGKPV QGVAYVRFGL LDEDGKKTFF RGLE SQTKL VNGQSHISLS KAEFQDALEK LNMGITDLQG LRLYVAAAII ESPGGEMEEA ELTSW YFVS SPFSLDLSKT KRHLVPGAPF LLQALVREMS GSPASGIPVK VSATVSSPGS VPEVQD IQQ NTDGSGQVSI PIIIPQTISE LQLSVSAGSP HPAIARLTVA APPSGGPGFL SIERPDS RP PRVGDTLNLN LRAVGSGATF SHYYYMILSR GQIVFMNREP KRTLTSVSVF VDHHLAPS F YFVAFYYHGD HPVANSLRVD VQAGACEGKL ELSVDGAKQY RNGESVKLHL ETDSLALVA LGALDTALYA AGSKSHKPLN MGKVFEAMNS YDLGCGPGGG DSALQVFQAA GLAFSDGDQW TLSRKRLSC PKEKTTRKKR NVNFQKAINE KLGQYASPTA KRCCQDGVTR LPMMRSCEQR A ARVQQPDC REPFLSCCQF AESLRKKSRD KGQAGLQRAL EILQEEDLID EDDIPVRSFF PE NWLWRVE TVDRFQILTL WLPDSLTTWE IHGLSLSKTK GLCVATPVQL RVFREFHLHL RLP MSVRRF EQLELRPVLY NYLDKNLTVS VHVSPVEGLC LAGGGGLAQQ VLVPAGSARP VAFS VVPTA AAAVSLKVVA RGSFEFPVGD AVSKVLQIEK EGAIHREELV YELNPLDHRG RTLEI PGNS DPNMIPDGDF NSYVRVTASD PLDTLGSEGA LSPGGVASLL RLPRGCGEQT MIYLAP TLA ASRYLDKTEQ WSTLPPETKD HAVDLIQKGY MRIQQFRKAD GSYAAWLSRD SSTWLTA FV LKVLSLAQEQ VGGSPEKLQE TSNWLLSQQQ ADGSFQDPCP VLDRSMQGGL VGNDETVA L TAFVTIALHH GLAVFQDEGA EPLKQRVEAS ISKANSFLGE KASAGLLGAH AAAITAYAL SLTKAPVDLL GVAHNNLMAM AQETGDNLYW GSVTGSQSNA VSPTPAPRNP SDPMPQAPAL WIETTAYAL LHLLLHEGKA EMADQASAWL TRQGSFQGGF RSTQDTVIAL DALSAYWIAS H TTEERGLN VTLSSTGRNG FKSHALQLNN RQIRGLEEEL QFSLGSKINV KVGGNSKGTL KV LRTYNVL DMKNTTCQDL QIEVTVKGHV EYTMEANEDY EDYEYDELPA KDDPDAPLQP VTP LQLFEG RRNRRRREAP KVVEEQESRV HYTVCIWRNG KVGLSGMAIA DVTLLSGFHA LRAD LEKLT SLSDRYVSHF ETEGPHVLLY FDSVPTSREC VGFEAVQEVP VGLVQPASAT LYDYY NPER RCSVFYGAPS KSRLLATLCS AEVCQCAEGK CPRQRRALER GLQDEDGYRM KFACYY PRV EYGFQVKVLR EDSRAAFRLF ETKITQVLHF TKDVKAAANQ MRNFLVRASC RLRLEPG KE YLIMGLDGAT YDLEGHPQYL LDSNSWIEEM PSERLCRSTR QRAACAQLND FLQEYGTQ G CQV |
-Macromolecule #2: Nanobody G3
| Macromolecule | Name: Nanobody G3 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQAGGSLRL SCAASESIFI NNMGWFRQAP GKERELVATI ARDYGPNYAD SAKGRFTITR DNAKNMYLQM NNLKTEDTGV YYCRVVVAGG YYYWGQGTQV TVSSHGSGLV PRGSGGGHHH HHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.3 Component:
Details: 1x PBS. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot 4.5 seconds, force 1. | |||||||||
| Details | Purified components were mixed before vitrification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 1658 / Average exposure time: 6.4 sec. / Average electron dose: 50.5 e/Å2 / Details: 32 frames of 0.2 seconds. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)