+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11852 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bovine Papillomavirus E1 DNA helicase-replication fork complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA / virus / helicase / replisome / DNA replication. / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA 3'-5' helicase / DNA helicase activity / DNA replication / host cell nucleus / ATP hydrolysis activity / DNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Bovine papillomavirus Bovine papillomavirus | |||||||||
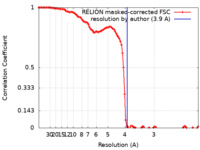
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Javed A / Major B | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Unwinding of a DNA replication fork by a hexameric viral helicase. Authors: Abid Javed / Balazs Major / Jonathan A Stead / Cyril M Sanders / Elena V Orlova /  Abstract: Hexameric helicases are motor proteins that unwind double-stranded DNA (dsDNA) during DNA replication but how they are optimised for strand separation is unclear. Here we present the cryo-EM ...Hexameric helicases are motor proteins that unwind double-stranded DNA (dsDNA) during DNA replication but how they are optimised for strand separation is unclear. Here we present the cryo-EM structure of the full-length E1 helicase from papillomavirus, revealing all arms of a bound DNA replication fork and their interactions with the helicase. The replication fork junction is located at the entrance to the helicase collar ring, that sits above the AAA + motor assembly. dsDNA is escorted to and the 5´ single-stranded DNA (ssDNA) away from the unwinding point by the E1 dsDNA origin binding domains. The 3´ ssDNA interacts with six spirally-arranged β-hairpins and their cyclical top-to-bottom movement pulls the ssDNA through the helicase. Pulling of the RF against the collar ring separates the base-pairs, while modelling of the conformational cycle suggest an accompanying movement of the collar ring has an auxiliary role, helping to make efficient use of ATP in duplex unwinding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11852.map.gz emd_11852.map.gz | 64.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11852-v30.xml emd-11852-v30.xml emd-11852.xml emd-11852.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11852_fsc.xml emd_11852_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11852.png emd_11852.png | 134.4 KB | ||
| Filedesc metadata |  emd-11852.cif.gz emd-11852.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11852 http://ftp.pdbj.org/pub/emdb/structures/EMD-11852 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11852 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11852 | HTTPS FTP |
-Related structure data
| Related structure data |  7apdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11852.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11852.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BPV E1 DNA helicase-replication fork complex
| Entire | Name: BPV E1 DNA helicase-replication fork complex |
|---|---|
| Components |
|
-Supramolecule #1: BPV E1 DNA helicase-replication fork complex
| Supramolecule | Name: BPV E1 DNA helicase-replication fork complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 413.4 KDa |
-Supramolecule #2: DNA replication fork
| Supramolecule | Name: DNA replication fork / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3-#4 Details: Consists of two strands and three regions: dsDNA, 5'ssDNA lagging strand and 3' ssDNA leading strand. |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
-Supramolecule #3: Full-length E1 helicase
| Supramolecule | Name: Full-length E1 helicase / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 Details: Composed of six subunits; Two subunits contain the Origin Binding domains (Chains G, H), all six subunits contain the helicase domain and the C-terminal tail (Chains A-F). |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
-Macromolecule #1: Replication protein E1
| Macromolecule | Name: Replication protein E1 / type: protein_or_peptide / ID: 1 / Details: OBD domains from subunits B and E. / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
| Molecular weight | Theoretical: 17.162084 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSRATVFKLG LFKSLFLCSF HDITRLFKND KTTNQQWVLA VFGLAEVFFE ASFELLKKQC SFLQMQKRSH EGGTCAVYLI CFNTAKSRE TVRNLMANML NVREECLMLQ PPKIRGLSAA LFWFKSSLSP ATLKHGALPE WIRAQTTLNA AAA UniProtKB: Replication protein E1 |
-Macromolecule #2: Replication protein E1
| Macromolecule | Name: Replication protein E1 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
| Molecular weight | Theoretical: 33.859172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TEKFDFGTMV QWAYDHKYAE ESKIAYEYAL AAGSDSNARA FLATNSQAKH VKDCATMVRH YLRAETQALS MPAYIKARCK LATGEGSWK SILTFFNYQN IELITFINAL KLWLKGIPKK NCLAFIGPPN TGKSMLCNSL IHFLGGSVLS FANHKSHFWL A SLADTRAA ...String: TEKFDFGTMV QWAYDHKYAE ESKIAYEYAL AAGSDSNARA FLATNSQAKH VKDCATMVRH YLRAETQALS MPAYIKARCK LATGEGSWK SILTFFNYQN IELITFINAL KLWLKGIPKK NCLAFIGPPN TGKSMLCNSL IHFLGGSVLS FANHKSHFWL A SLADTRAA LVDDATHACW RYFDTYLRNA LDGYPVSIDR KHKAAVQIKA PPLLVTSNID VQAEDRYLYL HSRVQTFRFE QP CTDESGE QPFNITDADW KSFFVRLWGR LDLIDEEEDS EEDGDSMRTF TCSARNTNAV D UniProtKB: Replication protein E1 |
-Macromolecule #3: DNA (40-MER)
| Macromolecule | Name: DNA (40-MER) / type: dna / ID: 3 / Details: 5'-3' ssDNA strand of the DNA replication fork. / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
| Molecular weight | Theoretical: 12.142779 KDa |
| Sequence | String: (DT)(DG)(DT)(DA)(DT)(DT)(DT)(DC)(DA)(DC) (DA)(DC)(DC)(DG)(DC)(DA)(DC)(DC)(DT)(DC) (DA)(DG)(DC)(DG)(DC)(DG)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) |
-Macromolecule #4: DNA (36-MER)
| Macromolecule | Name: DNA (36-MER) / type: dna / ID: 4 / Details: 3'-5' ssDNA strand of the DNA replication fork. / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Bovine papillomavirus Bovine papillomavirus |
| Molecular weight | Theoretical: 11.08009 KDa |
| Sequence | String: (DC)(DC)(DC)(DC)(DC)(DC)(DC)(DG)(DT)(DG) (DC)(DG)(DC)(DG)(DC)(DT)(DG)(DA)(DG)(DG) (DT)(DG)(DC)(DG)(DG)(DT)(DG)(DT)(DG) (DA)(DA)(DA)(DT)(DA)(DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV Details: 3 ul of sample was applied Lacey ultra-thin carbon film grids.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 95.0 K / Max: 98.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 12136 / Average exposure time: 3.0 sec. / Average electron dose: 50.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 102 / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-7apd: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)