[English] 日本語
 Yorodumi
Yorodumi- EMDB-11314: The shoulder domain of dynactin with underlying Arp1 filament subnits -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11314 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The shoulder domain of dynactin with underlying Arp1 filament subnits | |||||||||
 Map data Map data | Map is aligned using EMDA to EMD-11313. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
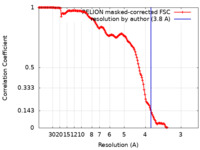
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Lau CK / Lacey SE / Carter AP | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end. Authors: Clinton K Lau / Francis J O'Reilly / Balaji Santhanam / Samuel E Lacey / Juri Rappsilber / Andrew P Carter /   Abstract: Dynactin is a 1.1 MDa complex that activates the molecular motor dynein for ultra-processive transport along microtubules. In order to do this, it forms a tripartite complex with dynein and a coiled- ...Dynactin is a 1.1 MDa complex that activates the molecular motor dynein for ultra-processive transport along microtubules. In order to do this, it forms a tripartite complex with dynein and a coiled-coil adaptor. Dynactin consists of an actin-related filament whose length is defined by its flexible shoulder domain. Despite previous cryo-EM structures, the molecular architecture of the shoulder and pointed end of the filament is still poorly understood due to the lack of high-resolution information in these regions. Here we combine multiple cryo-EM datasets and define precise masking strategies for particle signal subtraction and 3D classification. This overcomes domain flexibility and results in high-resolution maps into which we can build the shoulder and pointed end. The unique architecture of the shoulder securely houses the p150 subunit and positions the four identical p50 subunits in different conformations to bind dynactin's filament. The pointed end map allows us to build the first structure of p62 and reveals the molecular basis for cargo adaptor binding to different sites at the pointed end. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11314.map.gz emd_11314.map.gz | 251.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11314-v30.xml emd-11314-v30.xml emd-11314.xml emd-11314.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11314_fsc.xml emd_11314_fsc.xml | 15.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_11314.png emd_11314.png | 59.8 KB | ||
| Masks |  emd_11314_msk_1.map emd_11314_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Others |  emd_11314_additional_1.map.gz emd_11314_additional_1.map.gz emd_11314_additional_2.map.gz emd_11314_additional_2.map.gz emd_11314_additional_3.map.gz emd_11314_additional_3.map.gz emd_11314_half_map_1.map.gz emd_11314_half_map_1.map.gz emd_11314_half_map_2.map.gz emd_11314_half_map_2.map.gz | 283.2 MB 277.9 MB 569.9 KB 244 MB 244 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11314 http://ftp.pdbj.org/pub/emdb/structures/EMD-11314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11314 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11314 | HTTPS FTP |
-Validation report
| Summary document |  emd_11314_validation.pdf.gz emd_11314_validation.pdf.gz | 358.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11314_full_validation.pdf.gz emd_11314_full_validation.pdf.gz | 357.5 KB | Display | |
| Data in XML |  emd_11314_validation.xml.gz emd_11314_validation.xml.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11314 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11314 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11314 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11314 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11314.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11314.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map is aligned using EMDA to EMD-11313. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11314_msk_1.map emd_11314_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
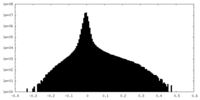
| Density Histograms |
-Additional map: Same map as main map, but before fitting using EMDA
| File | emd_11314_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Same map as main map, but before fitting using EMDA | ||||||||||||
| Projections & Slices |
| ||||||||||||
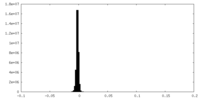
| Density Histograms |
-Additional map: Same map as main map, filtered to 6 Angstrom
| File | emd_11314_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Same map as main map, filtered to 6 Angstrom | ||||||||||||
| Projections & Slices |
| ||||||||||||
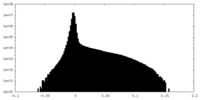
| Density Histograms |
-Additional map: Mask aligned on map before fitting
| File | emd_11314_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mask aligned on map before fitting | ||||||||||||
| Projections & Slices |
| ||||||||||||
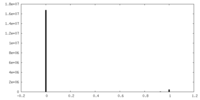
| Density Histograms |
-Half map: #2
| File | emd_11314_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11314_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dynactin complex
| Entire | Name: Dynactin complex |
|---|---|
| Components |
|
-Supramolecule #1: Dynactin complex
| Supramolecule | Name: Dynactin complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#17 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1 MDa |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)

























































 Processing
Processing Electron microscopy #1
Electron microscopy #1