[English] 日本語
 Yorodumi
Yorodumi- EMDB-11279: Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11279 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P/P tRNA | |||||||||
 Map data Map data | Combined map of five local-masked refined maps with sharpening and local-resolution filtering. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondrion / translation / closed nascent-polypeptide tunnel / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationrRNA import into mitochondrion / mitochondrial transcription / mitochondrial translational termination / Mitochondrial ribosome-associated quality control / mitochondrial translational elongation / mitochondrial ribosome assembly / Mitochondrial translation elongation / translation release factor activity, codon nonspecific / Mitochondrial translation initiation / Mitochondrial translation termination ...rRNA import into mitochondrion / mitochondrial transcription / mitochondrial translational termination / Mitochondrial ribosome-associated quality control / mitochondrial translational elongation / mitochondrial ribosome assembly / Mitochondrial translation elongation / translation release factor activity, codon nonspecific / Mitochondrial translation initiation / Mitochondrial translation termination / negative regulation of mitotic nuclear division / mitochondrial large ribosomal subunit / peptidyl-tRNA hydrolase / translation release factor activity / mitochondrial ribosome / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / mitochondrial small ribosomal subunit / peptidyl-tRNA hydrolase activity / mitochondrial translation / apoptotic mitochondrial changes / positive regulation of proteolysis / ribosomal small subunit binding / anatomical structure morphogenesis / RNA processing / Mitochondrial protein degradation / rescue of stalled cytosolic ribosome / cellular response to leukemia inhibitory factor / apoptotic signaling pathway / fibrillar center / cell junction / double-stranded RNA binding / regulation of translation / ribosomal small subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / endonuclease activity / nuclear membrane / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / tRNA binding / cell population proliferation / mitochondrial inner membrane / negative regulation of translation / rRNA binding / nuclear body / structural constituent of ribosome / ribosome / translation / mitochondrial matrix / intracellular membrane-bounded organelle / ribonucleoprotein complex / protein domain specific binding / nucleotide binding / mRNA binding / apoptotic process / positive regulation of DNA-templated transcription / GTP binding / nucleolus / mitochondrion / extracellular space / RNA binding / nucleoplasm / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
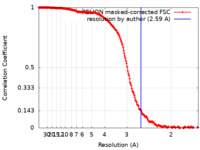
| Method | single particle reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||
 Authors Authors | Itoh Y / Andrell J | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Mechanism of membrane-tethered mitochondrial protein synthesis. Authors: Yuzuru Itoh / Juni Andréll / Austin Choi / Uwe Richter / Priyanka Maiti / Robert B Best / Antoni Barrientos / Brendan J Battersby / Alexey Amunts /     Abstract: Mitochondrial ribosomes (mitoribosomes) are tethered to the mitochondrial inner membrane to facilitate the cotranslational membrane insertion of the synthesized proteins. We report cryo-electron ...Mitochondrial ribosomes (mitoribosomes) are tethered to the mitochondrial inner membrane to facilitate the cotranslational membrane insertion of the synthesized proteins. We report cryo-electron microscopy structures of human mitoribosomes with nascent polypeptide, bound to the insertase oxidase assembly 1-like (OXA1L) through three distinct contact sites. OXA1L binding is correlated with a series of conformational changes in the mitoribosomal large subunit that catalyze the delivery of newly synthesized polypeptides. The mechanism relies on the folding of mL45 inside the exit tunnel, forming two specific constriction sites that would limit helix formation of the nascent chain. A gap is formed between the exit and the membrane, making the newly synthesized proteins accessible. Our data elucidate the basis by which mitoribosomes interact with the OXA1L insertase to couple protein synthesis and membrane delivery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11279.map.gz emd_11279.map.gz | 339.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11279-v30.xml emd-11279-v30.xml emd-11279.xml emd-11279.xml | 130.4 KB 130.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11279_fsc.xml emd_11279_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11279.png emd_11279.png | 81 KB | ||
| Masks |  emd_11279_msk_1.map emd_11279_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11279.cif.gz emd-11279.cif.gz | 23.5 KB | ||
| Others |  emd_11279_additional_1.map.gz emd_11279_additional_1.map.gz emd_11279_half_map_1.map.gz emd_11279_half_map_1.map.gz emd_11279_half_map_2.map.gz emd_11279_half_map_2.map.gz | 557.7 MB 486.5 MB 486.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11279 http://ftp.pdbj.org/pub/emdb/structures/EMD-11279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11279 | HTTPS FTP |
-Related structure data
| Related structure data |  6zm6MC  6zm5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11279.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11279.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined map of five local-masked refined maps with sharpening and local-resolution filtering. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11279_msk_1.map emd_11279_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
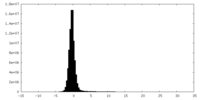
| Density Histograms |
-Additional map: Combined map of five local-masked refined maps without sharpening.
| File | emd_11279_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined map of five local-masked refined maps without sharpening. | ||||||||||||
| Projections & Slices |
| ||||||||||||
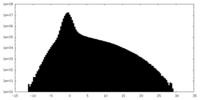
| Density Histograms |
-Half map: Local-masked refined map using the mask covering the LSU-core.
| File | emd_11279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-masked refined map using the mask covering the LSU-core. | ||||||||||||
| Projections & Slices |
| ||||||||||||
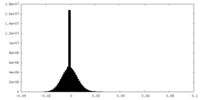
| Density Histograms |
-Half map: Local-masked refined map using the mask covering the LSU-core.
| File | emd_11279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-masked refined map using the mask covering the LSU-core. | ||||||||||||
| Projections & Slices |
| ||||||||||||
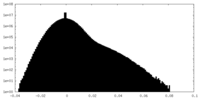
| Density Histograms |
- Sample components
Sample components
+Entire : Ribosome with mRNA and tRNAs
+Supramolecule #1: Ribosome with mRNA and tRNAs
+Supramolecule #2: Large subunit of ribosome
+Supramolecule #3: Small subunit of ribosome
+Macromolecule #1: 16S mitochondrial rRNA
+Macromolecule #2: mitochondrial tRNAVal
+Macromolecule #54: 12S mitochondrial rRNA
+Macromolecule #85: A-site tRNA
+Macromolecule #86: P-site tRNA
+Macromolecule #87: mRNA
+Macromolecule #3: 39S ribosomal protein L2, mitochondrial
+Macromolecule #4: 39S ribosomal protein L3, mitochondrial
+Macromolecule #5: 39S ribosomal protein L4, mitochondrial
+Macromolecule #6: 39S ribosomal protein L9, mitochondrial
+Macromolecule #7: 39S ribosomal protein L10, mitochondrial
+Macromolecule #8: 39S ribosomal protein L11, mitochondrial
+Macromolecule #9: 39S ribosomal protein L13, mitochondrial
+Macromolecule #10: 39S ribosomal protein L14, mitochondrial
+Macromolecule #11: 39S ribosomal protein L15, mitochondrial
+Macromolecule #12: 39S ribosomal protein L16, mitochondrial
+Macromolecule #13: 39S ribosomal protein L17, mitochondrial
+Macromolecule #14: 39S ribosomal protein L18, mitochondrial
+Macromolecule #15: 39S ribosomal protein L19, mitochondrial
+Macromolecule #16: 39S ribosomal protein L20, mitochondrial
+Macromolecule #17: 39S ribosomal protein L21, mitochondrial
+Macromolecule #18: 39S ribosomal protein L22, mitochondrial
+Macromolecule #19: 39S ribosomal protein L23, mitochondrial
+Macromolecule #20: 39S ribosomal protein L24, mitochondrial
+Macromolecule #21: 39S ribosomal protein L27, mitochondrial
+Macromolecule #22: 39S ribosomal protein L28, mitochondrial
+Macromolecule #23: 39S ribosomal protein L47, mitochondrial
+Macromolecule #24: 39S ribosomal protein L30, mitochondrial
+Macromolecule #25: 39S ribosomal protein L32, mitochondrial
+Macromolecule #26: 39S ribosomal protein L33, mitochondrial
+Macromolecule #27: 39S ribosomal protein L34, mitochondrial
+Macromolecule #28: 39S ribosomal protein L35, mitochondrial
+Macromolecule #29: 39S ribosomal protein L36, mitochondrial
+Macromolecule #30: 39S ribosomal protein L37, mitochondrial
+Macromolecule #31: 39S ribosomal protein L38, mitochondrial
+Macromolecule #32: 39S ribosomal protein L39, mitochondrial
+Macromolecule #33: 39S ribosomal protein L40, mitochondrial
+Macromolecule #34: 39S ribosomal protein L41, mitochondrial
+Macromolecule #35: 39S ribosomal protein L42, mitochondrial
+Macromolecule #36: 39S ribosomal protein L43, mitochondrial
+Macromolecule #37: 39S ribosomal protein L44, mitochondrial
+Macromolecule #38: 39S ribosomal protein L45, mitochondrial
+Macromolecule #39: 39S ribosomal protein L46, mitochondrial
+Macromolecule #40: 39S ribosomal protein L48, mitochondrial
+Macromolecule #41: 39S ribosomal protein L49, mitochondrial
+Macromolecule #42: 39S ribosomal protein L50, mitochondrial
+Macromolecule #43: 39S ribosomal protein L51, mitochondrial
+Macromolecule #44: 39S ribosomal protein L52, mitochondrial
+Macromolecule #45: 39S ribosomal protein L53, mitochondrial
+Macromolecule #46: 39S ribosomal protein L54, mitochondrial
+Macromolecule #47: 39S ribosomal protein L55, mitochondrial
+Macromolecule #48: Ribosomal protein 63, mitochondrial
+Macromolecule #49: Peptidyl-tRNA hydrolase ICT1, mitochondrial
+Macromolecule #50: Growth arrest and DNA damage-inducible proteins-interacting protein 1
+Macromolecule #51: 39S ribosomal protein S18a, mitochondrial
+Macromolecule #52: 39S ribosomal protein S30, mitochondrial
+Macromolecule #53: 39S ribosomal protein L12, mitochondrial
+Macromolecule #55: 28S ribosomal protein S2, mitochondrial
+Macromolecule #56: 28S ribosomal protein S24, mitochondrial
+Macromolecule #57: 28S ribosomal protein S5, mitochondrial
+Macromolecule #58: 28S ribosomal protein S6, mitochondrial
+Macromolecule #59: 28S ribosomal protein S7, mitochondrial
+Macromolecule #60: 28S ribosomal protein S9, mitochondrial
+Macromolecule #61: 28S ribosomal protein S10, mitochondrial
+Macromolecule #62: 28S ribosomal protein S11, mitochondrial
+Macromolecule #63: 28S ribosomal protein S12, mitochondrial
+Macromolecule #64: 28S ribosomal protein S14, mitochondrial
+Macromolecule #65: 28S ribosomal protein S15, mitochondrial
+Macromolecule #66: 28S ribosomal protein S16, mitochondrial
+Macromolecule #67: 28S ribosomal protein S17, mitochondrial
+Macromolecule #68: 28S ribosomal protein S18b, mitochondrial
+Macromolecule #69: 28S ribosomal protein S18c, mitochondrial
+Macromolecule #70: 28S ribosomal protein S21, mitochondrial
+Macromolecule #71: 28S ribosomal protein S22, mitochondrial
+Macromolecule #72: 28S ribosomal protein S23, mitochondrial
+Macromolecule #73: 28S ribosomal protein S25, mitochondrial
+Macromolecule #74: 28S ribosomal protein S26, mitochondrial
+Macromolecule #75: 28S ribosomal protein S27, mitochondrial
+Macromolecule #76: 28S ribosomal protein S28, mitochondrial
+Macromolecule #77: 28S ribosomal protein S29, mitochondrial
+Macromolecule #78: 28S ribosomal protein S31, mitochondrial
+Macromolecule #79: 28S ribosomal protein S33, mitochondrial
+Macromolecule #80: 28S ribosomal protein S34, mitochondrial
+Macromolecule #81: 28S ribosomal protein S35, mitochondrial
+Macromolecule #82: Coiled-coil-helix-coiled-coil-helix domain-containing protein 1
+Macromolecule #83: Aurora kinase A-interacting protein
+Macromolecule #84: Pentatricopeptide repeat domain-containing protein 3, mitochondrial
+Macromolecule #88: MAGNESIUM ION
+Macromolecule #89: POTASSIUM ION
+Macromolecule #90: ZINC ION
+Macromolecule #91: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #92: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #93: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #94: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 3 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-20 / Number grids imaged: 2 / Number real images: 19655 / Average exposure time: 4.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.6 µm / Calibrated defocus min: 0.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 44 / Target criteria: Correlation coefficient | ||||||||
| Output model |  PDB-6zm6: |
 Movie
Movie Controller
Controller








































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)