[English] 日本語
 Yorodumi
Yorodumi- EMDB-1109: Structural insights into the activity of enhancer-binding proteins. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1109 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into the activity of enhancer-binding proteins. | |||||||||
 Map data Map data | this is a volume map of PspF in complex with sigma54 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | RNA polymerase sigma factor 54 / AAA+ ATPase domain Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Rappas M / Schumacher J / Beuron F / Niwa H / Bordes P / Wigneshweraraj S / Keetch CA / Robinson CV / Buck M / Zhang X | |||||||||
 Citation Citation |  Journal: Science / Year: 2005 Journal: Science / Year: 2005Title: Structural insights into the activity of enhancer-binding proteins. Authors: Mathieu Rappas / Jorg Schumacher / Fabienne Beuron / Hajime Niwa / Patricia Bordes / Sivaramesh Wigneshweraraj / Catherine A Keetch / Carol V Robinson / Martin Buck / Xiaodong Zhang /  Abstract: Activators of bacterial sigma54-RNA polymerase holoenzyme are mechanochemical proteins that use adenosine triphosphate (ATP) hydrolysis to activate transcription. We have determined by cryogenic ...Activators of bacterial sigma54-RNA polymerase holoenzyme are mechanochemical proteins that use adenosine triphosphate (ATP) hydrolysis to activate transcription. We have determined by cryogenic electron microscopy (cryo-EM) a 20 angstrom resolution structure of an activator, phage shock protein F [PspF(1-275)], which is bound to an ATP transition state analog in complex with its basal factor, sigma54. By fitting the crystal structure of PspF(1-275) at 1.75 angstroms into the EM map, we identified two loops involved in binding sigma54. Comparing enhancer-binding structures in different nucleotide states and mutational analysis led us to propose nucleotide-dependent conformational changes that free the loops for association with sigma54. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1109.map.gz emd_1109.map.gz | 621.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1109-v30.xml emd-1109-v30.xml emd-1109.xml emd-1109.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  1109.gif 1109.gif | 23.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1109 http://ftp.pdbj.org/pub/emdb/structures/EMD-1109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1109 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1109 | HTTPS FTP |
-Validation report
| Summary document |  emd_1109_validation.pdf.gz emd_1109_validation.pdf.gz | 187.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1109_full_validation.pdf.gz emd_1109_full_validation.pdf.gz | 186.2 KB | Display | |
| Data in XML |  emd_1109_validation.xml.gz emd_1109_validation.xml.gz | 5.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1109 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1109 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1109 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1109.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1109.map.gz / Format: CCP4 / Size: 12.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | this is a volume map of PspF in complex with sigma54 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PspF AAA domain
| Entire | Name: PspF AAA domain |
|---|---|
| Components |
|
-Supramolecule #1000: PspF AAA domain
| Supramolecule | Name: PspF AAA domain / type: sample / ID: 1000 Oligomeric state: one hexamer of PspF binds to a monomer of sigma54 Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 240 KDa / Theoretical: 240 KDa / Method: mass spec |
-Macromolecule #1: PspF AAA domain
| Macromolecule | Name: PspF AAA domain / type: protein_or_peptide / ID: 1 / Name.synonym: pspf AAA domain / Number of copies: 6 / Oligomeric state: hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 31 KDa / Theoretical: 31 KDa |
| Recombinant expression | Organism: Escherichia coli B834 / Recombinant plasmid: pET28 bplus |
| Sequence | InterPro: AAA+ ATPase domain |
-Macromolecule #2: sigma54
| Macromolecule | Name: sigma54 / type: protein_or_peptide / ID: 2 / Name.synonym: sigma54 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 54 KDa / Theoretical: 54 KDa |
| Recombinant expression | Organism: B834 / Recombinant plasmid: pET28 bplus |
| Sequence | InterPro: RNA polymerase sigma factor 54 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10 mM Tris HCl, pH8, 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 5% glycerol |
| Staining | Type: NEGATIVE Details: Grids with native sample quench frozen in liquid ethane cooled at -186 C |
| Grid | Details: Holey carbon grids from Agar |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 87.15 K / Timed resolved state: vitrified 30msec after / Method: blot for 2 seconds |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG/UT |
|---|---|
| Temperature | Average: 103 K |
| Details | weak beam illumination |
| Date | Apr 4, 2002 |
| Image recording | Category: CCD / Film or detector model: KODAK SO-163 FILM / Digitization - Sampling interval: 2 µm / Number real images: 9 / Average electron dose: 20 e/Å2 / Bits/pixel: 14 |
| Electron beam | Acceleration voltage: 160 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 1.2 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 3 SIGMA CUT-OFF / Software - Name: IMAGIC-5 Details: Final maps were calculate from 123 class averages from one dataset Number images used: 3895 |
| Final two d classification | Number classes: 123 |
-Atomic model buiding 1
| Details | Protocol: Cross correlation coefficient between projections of fitted model and those from the EM reconstruction. The monomer was manually fitted using the program "O"; the model was p6 symmetrised and individually subunits were readjusted manually using "O" |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera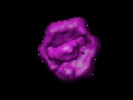







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)