[English] 日本語
 Yorodumi
Yorodumi- EMDB-1104: Global conformational rearrangements during the activation of the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1104 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3. | |||||||||
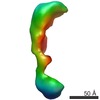
 Map data Map data | 3D reconstruction of phosphorylated (active) form of Vav3 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | small GTPase-mediated signal transduction Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Llorca O / Arias-Palomo E / Zugaza JL / Bustelo XR | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2005 Journal: EMBO J / Year: 2005Title: Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3. Authors: Oscar Llorca / Ernesto Arias-Palomo / José L Zugaza / Xosé R Bustelo /  Abstract: Activation of Rho/Rac GTPases during cell signaling requires the participation of GDP/GTP exchange factors of the Dbl family. Although the structure of the catalytic core of Dbl proteins has been ...Activation of Rho/Rac GTPases during cell signaling requires the participation of GDP/GTP exchange factors of the Dbl family. Although the structure of the catalytic core of Dbl proteins has been established recently, the molecular changes that the full-length proteins experience during normal or oncogenic conditions of stimulation are still unknown. Here, we have used single-particle electron microscopy to solve the structures of the inactive (unphosphorylated), active (phosphorylated), and constitutively active (N-terminally deleted) versions of the exchange factor Vav3. Comparison of these forms has revealed the interdomain interactions maintaining the inactive Vav3 state and the dynamic changes that the overall Vav3 structure undergoes upon tyrosine phosphorylation. We have also found that the conformations of phosphorylated Vav3 and N-terminally deleted Vav3 are distinct, indicating that the acquisition of constitutive activity by exchange factors is structurally more complex than the mere elimination of inhibitory interactions between structural domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1104.map.gz emd_1104.map.gz | 69.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1104-v30.xml emd-1104-v30.xml emd-1104.xml emd-1104.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  1104.gif 1104.gif emd_1104.tif emd_1104.tif | 3.1 KB 179.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1104 http://ftp.pdbj.org/pub/emdb/structures/EMD-1104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1104 | HTTPS FTP |
-Validation report
| Summary document |  emd_1104_validation.pdf.gz emd_1104_validation.pdf.gz | 189.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1104_full_validation.pdf.gz emd_1104_full_validation.pdf.gz | 188.9 KB | Display | |
| Data in XML |  emd_1104_validation.xml.gz emd_1104_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1104 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1104.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1104.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of phosphorylated (active) form of Vav3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Phosphorylated and active form of Vav3
| Entire | Name: Phosphorylated and active form of Vav3 |
|---|---|
| Components |
|
-Supramolecule #1000: Phosphorylated and active form of Vav3
| Supramolecule | Name: Phosphorylated and active form of Vav3 / type: sample / ID: 1000 / Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 98 KDa / Method: from its sequence |
-Macromolecule #1: phosphorylated vav3
| Macromolecule | Name: phosphorylated vav3 / type: protein_or_peptide / ID: 1 / Name.synonym: pY-vav3 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: baculovirus-infected S. frugiperda cells / Location in cell: cytoplasm Homo sapiens (human) / synonym: Human / Cell: baculovirus-infected S. frugiperda cells / Location in cell: cytoplasm |
| Molecular weight | Experimental: 98 KDa |
| Recombinant expression | Organism: baculovirus-infected S. frugiperda cells |
| Sequence | GO: small GTPase-mediated signal transduction |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM Tris-Hcl, 100 mM NaCl |
| Staining | Type: NEGATIVE Details: Glow-dischrged carbon-coated grids and negatively stained with 1% w/v uranyl acetate |
| Grid | Details: 400 mesh copper-rhodium grids |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Temperature | Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: correction with CCD camera |
| Details | Electron microscope:JEOL JEM-1230, 120kV. Specimen holder: JEOL type M:207EM-11020. |
| Date | Nov 14, 2003 |
| Image recording | Category: FILM / Film or detector model: KODAK 4489 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 10 µm / Details: Digitiser used: Minolta Dimage Scan Multi Pro / Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 3867 |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)


























