[English] 日本語
 Yorodumi
Yorodumi- EMDB-10896: First cryo-ET / subtomogram averaging of primary cilia microtubul... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10896 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
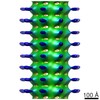
| Title | First cryo-ET / subtomogram averaging of primary cilia microtubules show that A-tubules are decorated by EB1 | |||||||||
 Map data Map data | Subtomogram average of EB1 decorated A-microtubule from MDCKII primary cilia axonemes | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 18.56 Å | |||||||||
 Authors Authors | Kiesel P / Alvarez Viar G / Tsoy N / Maraspini R / Honigmann A / Pigino G | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Authors: Petra Kiesel / Gonzalo Alvarez Viar / Nikolai Tsoy / Riccardo Maraspini / Peter Gorilak / Vladimir Varga / Alf Honigmann / Gaia Pigino /    Abstract: Primary cilia are microtubule-based organelles that are important for signaling and sensing in eukaryotic cells. Unlike the thoroughly studied motile cilia, the three-dimensional architecture and ...Primary cilia are microtubule-based organelles that are important for signaling and sensing in eukaryotic cells. Unlike the thoroughly studied motile cilia, the three-dimensional architecture and molecular composition of primary cilia are largely unexplored. Yet, studying these aspects is necessary to understand how primary cilia function in health and disease. We developed an enabling method for investigating the structure of primary cilia isolated from MDCK-II cells at molecular resolution by cryo-electron tomography. We show that the textbook '9 + 0' arrangement of microtubule doublets is only present at the primary cilium base. A few microns out, the architecture changes into an unstructured bundle of EB1-decorated microtubules and actin filaments, putting an end to a long debate on the presence or absence of actin filaments in primary cilia. Our work provides a plethora of insights into the molecular structure of primary cilia and offers a methodological framework to study these important organelles. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: The molecular structure of primary cilia revealed by cryo-electron tomography Authors: Kiesel P / Alvarez Viar G / Tsoy N / Maraspini R / Honigmann A / Pigino G | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10896.map.gz emd_10896.map.gz | 2.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10896-v30.xml emd-10896-v30.xml emd-10896.xml emd-10896.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10896.png emd_10896.png | 101.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10896 http://ftp.pdbj.org/pub/emdb/structures/EMD-10896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10896 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10896.map.gz / Format: CCP4 / Size: 3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10896.map.gz / Format: CCP4 / Size: 3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average of EB1 decorated A-microtubule from MDCKII primary cilia axonemes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.711 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Subtomogram average of EB1 decorated A-microtubule from cryo-tomo...
| Entire | Name: Subtomogram average of EB1 decorated A-microtubule from cryo-tomograms of MDCKII primary cilia |
|---|---|
| Components |
|
-Supramolecule #1: Subtomogram average of EB1 decorated A-microtubule from cryo-tomo...
| Supramolecule | Name: Subtomogram average of EB1 decorated A-microtubule from cryo-tomograms of MDCKII primary cilia type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.25 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















