+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10890 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-electron density map of the P140-P110 heterodimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P110/P140 Nap Mycoplasma genitalium surface adhesin / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationadhesion of symbiont to microvasculature / cell adhesion / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycoplasma genitalium G37 (bacteria) / Mycoplasma genitalium G37 (bacteria) /  Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) | |||||||||
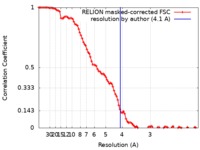
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Scheffer MP / Aparicio D | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure and mechanism of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. Authors: David Aparicio / Margot P Scheffer / Marina Marcos-Silva / David Vizarraga / Lasse Sprankel / Mercè Ratera / Miriam S Weber / Anja Seybert / Sergi Torres-Puig / Luis Gonzalez-Gonzalez / ...Authors: David Aparicio / Margot P Scheffer / Marina Marcos-Silva / David Vizarraga / Lasse Sprankel / Mercè Ratera / Miriam S Weber / Anja Seybert / Sergi Torres-Puig / Luis Gonzalez-Gonzalez / Julian Reitz / Enrique Querol / Jaume Piñol / Oscar Q Pich / Ignacio Fita / Achilleas S Frangakis /   Abstract: Mycoplasma genitalium is a human pathogen adhering to host target epithelial cells and causing urethritis, cervicitis and pelvic inflammatory disease. Essential for infectivity is a transmembrane ...Mycoplasma genitalium is a human pathogen adhering to host target epithelial cells and causing urethritis, cervicitis and pelvic inflammatory disease. Essential for infectivity is a transmembrane adhesion complex called Nap comprising proteins P110 and P140. Here we report the crystal structure of P140 both alone and in complex with the N-terminal domain of P110. By cryo-electron microscopy (cryo-EM) and tomography (cryo-ET) we find closed and open Nap conformations, determined at 9.8 and 15 Å, respectively. Both crystal structures and the cryo-EM structure are found in a closed conformation, where the sialic acid binding site in P110 is occluded. By contrast, the cryo-ET structure shows an open conformation, where the binding site is accessible. Structural information, in combination with functional studies, suggests a mechanism for attachment and release of M. genitalium to and from the host cell receptor, in which Nap conformations alternate to sustain motility and guarantee infectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10890.map.gz emd_10890.map.gz | 47.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10890-v30.xml emd-10890-v30.xml emd-10890.xml emd-10890.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10890_fsc.xml emd_10890_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10890.png emd_10890.png | 193.7 KB | ||
| Filedesc metadata |  emd-10890.cif.gz emd-10890.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10890 http://ftp.pdbj.org/pub/emdb/structures/EMD-10890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10890 | HTTPS FTP |
-Related structure data
| Related structure data |  6yrkMC  6rutC  6s3uC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10890.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10890.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : P140-P110 heterodimer
| Entire | Name: P140-P110 heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: P140-P110 heterodimer
| Supramolecule | Name: P140-P110 heterodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) |
-Macromolecule #1: Mgp-operon protein 3
| Macromolecule | Name: Mgp-operon protein 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) |
| Molecular weight | Theoretical: 84.816969 KDa |
| Recombinant expression | Organism:  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) |
| Sequence | String: TFLVKEDSKN VTAYTPFATP ITDSKSDLVS LAQLDSSYQI ADQTIHNTNL FVLFKSRDVK VKYESSGSNN ISFDSTSQGE KPSYVVEFT NSTGIKWTMV KKYQLDVPNV SSDMNQVLKN LILEQPLTKY TLNSSLAKEK GKTQREVHLG SGQANQWTSQ R NQHDLNNN ...String: TFLVKEDSKN VTAYTPFATP ITDSKSDLVS LAQLDSSYQI ADQTIHNTNL FVLFKSRDVK VKYESSGSNN ISFDSTSQGE KPSYVVEFT NSTGIKWTMV KKYQLDVPNV SSDMNQVLKN LILEQPLTKY TLNSSLAKEK GKTQREVHLG SGQANQWTSQ R NQHDLNNN PSPNASTGFK LTTGNAYRKL SESWPIYEPI DGTKQGKGKD SSGWSSTEEN EAKNDAPSVS SGTFNKYLNT KQ ALESIGI LFDDQTPRNV ITQLYYASTS KLAVTNNHIV VMGNSFLPSM WYWVVERSAQ ENASNKPTWF ANTNLDWGED KQK QFVENQ LGYKETTSTN SHNFHSKSFT QPAYLISGID SVNDQIIFSG FKAGSVGYDS SSSSSSSTKD QALAWSTTTS LDSK TGYKD LVTNDTGLNG PINGSFSIQD TFSFVVPYSG NHTNNGTTGP IKTAYPVKKD QKSTVKINSL INATPLNSYG DEGIG VFDA LGLNYNFKSN QERLPSRTDQ IFVYGIVSPN ELRSAKSSAD STGSDTKVNW SNTQSRYLPV PYNYSEGIID ADGFKR PEN RGASVTTFSG LKSIAPDGFA NSIANFSVGL KAGIDPNPVM SGKKANYGAV VLTRGGVVRL NFNPGNDSLL STTDNNI AP ISFSFTPFTA AESAVDLTTF KEVTYNQESG LWSYIFDSSL KPSHDGKQTP VTDNMGFSVI TVSRTGIELN QDQATTTL D VAPSALAVQS GIQSTTQTLT GVLPLSEEFS AVIAKDSDQN KIDIYKNNNG LFEIDT UniProtKB: Mgp-operon protein 3 |
-Macromolecule #2: Adhesin P1
| Macromolecule | Name: Adhesin P1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) (bacteria) |
| Molecular weight | Theoretical: 128.447648 KDa |
| Recombinant expression | Organism:  Mycoplasma genitalium G37 (bacteria) Mycoplasma genitalium G37 (bacteria) |
| Sequence | String: QAVDETLTPW TWNNNNFSSL KITGENPGSF GLVRSQNDNL NISSVTKDNL KYLNAVEKYL DGQQNFAIRR YDNNGRALYD INLAKMENP STVQRGLNGE PIFDPFKGFG LTGNAPTDWN EIKGKVPVEV VQSPHSPNLY FVLLVPKVAL EYHNLNNQVV K ESLEVSSF ...String: QAVDETLTPW TWNNNNFSSL KITGENPGSF GLVRSQNDNL NISSVTKDNL KYLNAVEKYL DGQQNFAIRR YDNNGRALYD INLAKMENP STVQRGLNGE PIFDPFKGFG LTGNAPTDWN EIKGKVPVEV VQSPHSPNLY FVLLVPKVAL EYHNLNNQVV K ESLEVSSF NPTQRLQKDS PVKDSSKQGE KLSETTASSM SSGMATSTRA KALKVEVERG SQSDSLLKND FAKKPLKHKN SS GEVKLEA EKEFTEAWKP LLTTDQIARE KGMGATVVSF YDAPYSENHT AFGLVDHIDP KKMVENYPPS WKTPKWNHHG IWD YNARNL LLQTTGFFNP RRHPEWFDEG QAKADNTSPG FKVGDTDHKK DGFKKNSSSP IALPFEAYFA NIGNMVAIGN SVFI FGGNG HATKMFTTNP LSIGVFRIKY TDNFSKSSVT GWPYAVLFGG LINPQTNGLK DLPLGTNRWF EYVPRMAVSG VKWVG NQLV LAGTLTMGDT ATVPRLKYDQ LEKHLNLVAQ GQGLLREDLQ IFTPYGWANR PDIPVGAWLQ DEMGSKFGPH YFLNNP DIQ DNVNNDTVEA LISSYKNTDK LKHVYPYRYS GLYAWQLFNW SNKLTNTPLS ANFVNENSYA PNSLFAAILN EDLLTGL SD KIFYGKENEF AENEADRFNQ LLSLNPNPNT NWARYLNVVQ RFTTGPNLDS STFDQFLDFL PWIGNGKPFS NSPSASTP L PTFSNINVGV KSMITQHLNK ENTRWVFIPN FSPDIWTGAG YRVQSANQKN GIPFEQVKPS NNSTPFDPNS DDNKVTPSG GSSKPTTYPA LPNSISPTSD WINALTFTNK NNPQRNQLLL RSLLGTIPVL INKSGDSNDQ FNKDSEQKWD KTETNEGNLP GFGEVNGLY NAALLHTYGF FGTNTNSTDP KIGFKADSSS SSSSTLVGSG LNWTSQDVGN LVVINDTSFG FQLGGWFITF T DFIRPRTG YLGITLSSLQ DQTIIWADQP WTSFKGSYLD SDGTPKSLWD PTALKSLPNN PTLSPSFQLY QPNKVKAYQT TN TYNKLIE PVDATSAATN MTSLLKLLTT KNIKAKLGKG TASNNGGGVS QTINTITTTG NISEGLKEET SIQAETLKKF FDS KQNNKS EIGIGDSTFT KMDGKLTGVV STPLVNLING QGAT UniProtKB: Adhesin P1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 7.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-6yrk: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)