[English] 日本語
 Yorodumi
Yorodumi- EMDB-10771: Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10771 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | |||||||||
 Map data Map data | engineered glycolyl-CoA carboxylase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | biotin dependent / ATP dependent / glycolyl-CoA / heterododecamer / enzyme engineering / CO2 fixation / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpropionyl-CoA carboxylase / propionyl-CoA carboxylase activity / acetyl-CoA carboxylase complex / acetyl-CoA carboxylase activity / carbon fixation / lipid catabolic process / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.96 Å | |||||||||
 Authors Authors | Schuller JM / Schuller SK | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Catal / Year: 2021 Journal: Nat Catal / Year: 2021Title: A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation Authors: Scheffen M / Marchal DG / Beneyton T / Schuller SK / Klose M / Diehl C / Lehmann J / Pfister P / Carrillo M / He H / Aslan S / Cortina NS / Claus P / Bollschweiler D / Baret JC / Schuller JM ...Authors: Scheffen M / Marchal DG / Beneyton T / Schuller SK / Klose M / Diehl C / Lehmann J / Pfister P / Carrillo M / He H / Aslan S / Cortina NS / Claus P / Bollschweiler D / Baret JC / Schuller JM / Zarzycki J / Bar-Even A / Erb TJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10771.map.gz emd_10771.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10771-v30.xml emd-10771-v30.xml emd-10771.xml emd-10771.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10771.png emd_10771.png | 221.7 KB | ||
| Filedesc metadata |  emd-10771.cif.gz emd-10771.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10771 http://ftp.pdbj.org/pub/emdb/structures/EMD-10771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10771 | HTTPS FTP |
-Related structure data
| Related structure data |  6ybqMC  6ybpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10771.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10771.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | engineered glycolyl-CoA carboxylase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
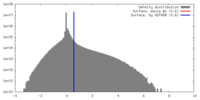
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Engineered glycolyl-CoA carboxylase with bound CoA
| Entire | Name: Engineered glycolyl-CoA carboxylase with bound CoA |
|---|---|
| Components |
|
-Supramolecule #1: Engineered glycolyl-CoA carboxylase with bound CoA
| Supramolecule | Name: Engineered glycolyl-CoA carboxylase with bound CoA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) |
| Molecular weight | Theoretical: 767 KDa |
-Macromolecule #1: Propionyl-CoA carboxylase beta chain
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) |
| Molecular weight | Theoretical: 55.963926 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKDILEKLEE RRAQARLGGG EKRLEAQHKR GKLTARERIE LLLDHGSFEE FDMFVQHRST DFGMEKQKIP GDGVVTGWGT VNGRTVFLF SKDFTVFGGS SSEAHAAKIV KVQDMALKMR APIIGIFDAG GARIQEGVAA LGGHGEVFRR NVAASGVIPQ I SVIMGPCA ...String: MKDILEKLEE RRAQARLGGG EKRLEAQHKR GKLTARERIE LLLDHGSFEE FDMFVQHRST DFGMEKQKIP GDGVVTGWGT VNGRTVFLF SKDFTVFGGS SSEAHAAKIV KVQDMALKMR APIIGIFDAG GARIQEGVAA LGGHGEVFRR NVAASGVIPQ I SVIMGPCA GGDVYSPAMT DFIFMVRDTS YMFVTGPDVV KTVTNEVVTA EELGGAKVHT SKSSIADGSF ENDVEAILQI RR LLDFLPA NNIEGVPEIE SFDDVNRLDK SLDTLIPDNP NKPYDMGELI RRVVDEGDFF EIQAAYARNI ITGFGRVEGR TVG FVANQP LVLAGVLDSD ASRKAARFVR FCNAFSIPIV TFVDVPGFLP GTAQEYGGLI KHGAKLLFAY SQATVPLVTI ITRK AFGGA YIVMASKHVG ADLNYAWPTA QIAVMGAKGA VEIIFRAEIG DADKVAERTK EYEDRFLSPF VAAERGYIDE VIMPH STRK RIARALGMLR TKEMEQPRKK HDNIPL UniProtKB: Propionyl-CoA carboxylase beta chain |
-Macromolecule #2: Propionyl-CoA carboxylase alpha subunit
| Macromolecule | Name: Propionyl-CoA carboxylase alpha subunit / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) Methylorubrum extorquens (strain ATCC 14718 / DSM 1338 / JCM 2805 / NCIMB 9133 / AM1) (bacteria) |
| Molecular weight | Theoretical: 71.986961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFDKILIANR GEIACRIIKT AQKMGIKTVA VYSDADRDAV HVAMADEAVH IGPAPAAQSY LLIEKIIDAC KQTGAQAVHP GYGFLSERE SFPKALAEAG IVFIGPNPGA IAAMGDKIES KKAAAAAEVS TVPGFLGVIE SPEHAVTIAD EIGYPVMIKA S AGGGGKGM ...String: MFDKILIANR GEIACRIIKT AQKMGIKTVA VYSDADRDAV HVAMADEAVH IGPAPAAQSY LLIEKIIDAC KQTGAQAVHP GYGFLSERE SFPKALAEAG IVFIGPNPGA IAAMGDKIES KKAAAAAEVS TVPGFLGVIE SPEHAVTIAD EIGYPVMIKA S AGGGGKGM RIAESADEVA EGFARAKSEA SSSFGDDRVF VEKFITDPRH IEIQVIGDKH GNVIYLGERE CSIQRRNQKV IE EAPSPLL DEETRRKMGE QAVALAKAVN YDSAGTVEFV AGQDKSFYFL EMNTRLQVEH PVTEMITGLD LVELMIRVAA GEK LPLSQD QVKLDGWAVE SRVYAEDPTR NFLPSIGRLT TYQPPEEGPL GGAIVRNDTG VEEGGEIAIH YDPMIAKLVT WAPT RLEAI EAQATALDAF AIEGIRHNIP FLATLMAHPR WRDGRLSTGF IKEEFPEGFI APEPEGPVAH RLAAVAAAID HKLNI RKRG ISGQMRDPSL LTFQRERVVV LSGQRFNVTV DPDGDDLLVT FDDGTTAPVR SAWRPGAPVW SGTVGDQSVA IQVRPL LNG VFLQHAGAAA EARVFTRREA ELADLMPVKE NAGSGKQLLC PMPGLVKQIM VSEGQEVKNG EPLAIVEAMK MENVLRA ER DGTISKIAAK EGDSLAVDAV ILEFA UniProtKB: propionyl-CoA carboxylase |
-Macromolecule #3: COENZYME A
| Macromolecule | Name: COENZYME A / type: ligand / ID: 3 / Number of copies: 6 / Formula: COA |
|---|---|
| Molecular weight | Theoretical: 767.534 Da |
| Chemical component information |  ChemComp-COA: |
-Macromolecule #4: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL
| Macromolecule | Name: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL type: ligand / ID: 4 / Number of copies: 6 / Formula: BTI |
|---|---|
| Molecular weight | Theoretical: 228.311 Da |
| Chemical component information |  ChemComp-BTI: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 871 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 1.96 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 2181317 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)