[English] 日本語
 Yorodumi
Yorodumi- EMDB-10648: CryoEM structure of the narrow type IV pilus (PilA5) from Thermus... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10648 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the narrow type IV pilus (PilA5) from Thermus thermophilus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type IV pilin glycosylation twitching motility / CELL ADHESION | |||||||||
| Function / homology | : / Prokaryotic N-terminal methylation site. / Prokaryotic N-terminal methylation motif / Prokaryotic N-terminal methylation site / Pilin-like / cell outer membrane / periplasmic space / plasma membrane / Type IV narrow pilus major component PilA5 Function and homology information Function and homology information | |||||||||
| Biological species |   Thermus thermophilus HB27 (bacteria) / Thermus thermophilus HB27 (bacteria) /   Thermus thermophilus (strain HB27 / ATCC BAA-163 / DSM 7039) (bacteria) Thermus thermophilus (strain HB27 / ATCC BAA-163 / DSM 7039) (bacteria) | |||||||||
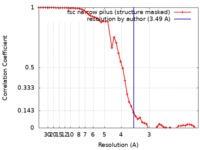
| Method | helical reconstruction / cryo EM / Resolution: 3.49 Å | |||||||||
 Authors Authors | Neuhaus A / Gold VAM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Cryo-electron microscopy reveals two distinct type IV pili assembled by the same bacterium. Authors: Alexander Neuhaus / Muniyandi Selvaraj / Ralf Salzer / Julian D Langer / Kerstin Kruse / Lennart Kirchner / Kelly Sanders / Bertram Daum / Beate Averhoff / Vicki A M Gold /    Abstract: Type IV pili are flexible filaments on the surface of bacteria, consisting of a helical assembly of pilin proteins. They are involved in bacterial motility (twitching), surface adhesion, biofilm ...Type IV pili are flexible filaments on the surface of bacteria, consisting of a helical assembly of pilin proteins. They are involved in bacterial motility (twitching), surface adhesion, biofilm formation and DNA uptake (natural transformation). Here, we use cryo-electron microscopy and mass spectrometry to show that the bacterium Thermus thermophilus produces two forms of type IV pilus ('wide' and 'narrow'), differing in structure and protein composition. Wide pili are composed of the major pilin PilA4, while narrow pili are composed of a so-far uncharacterized pilin which we name PilA5. Functional experiments indicate that PilA4 is required for natural transformation, while PilA5 is important for twitching motility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10648.map.gz emd_10648.map.gz | 361.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10648-v30.xml emd-10648-v30.xml emd-10648.xml emd-10648.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10648_fsc.xml emd_10648_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10648.png emd_10648.png | 118 KB | ||
| Filedesc metadata |  emd-10648.cif.gz emd-10648.cif.gz | 4.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10648 http://ftp.pdbj.org/pub/emdb/structures/EMD-10648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10648 | HTTPS FTP |
-Validation report
| Summary document |  emd_10648_validation.pdf.gz emd_10648_validation.pdf.gz | 447.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10648_full_validation.pdf.gz emd_10648_full_validation.pdf.gz | 447.3 KB | Display | |
| Data in XML |  emd_10648_validation.xml.gz emd_10648_validation.xml.gz | 8.6 KB | Display | |
| Data in CIF |  emd_10648_validation.cif.gz emd_10648_validation.cif.gz | 10.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10648 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10648 | HTTPS FTP |
-Related structure data
| Related structure data |  6xxeMC  6xxdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10648.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10648.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Thermus thermophilus narrow pilus
| Entire | Name: Thermus thermophilus narrow pilus |
|---|---|
| Components |
|
-Supramolecule #1: Thermus thermophilus narrow pilus
| Supramolecule | Name: Thermus thermophilus narrow pilus / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB27 (bacteria) Thermus thermophilus HB27 (bacteria) |
-Macromolecule #1: Uncharacterized protein
| Macromolecule | Name: Uncharacterized protein / type: protein_or_peptide / ID: 1 Details: Sequence corresponds to the mature protein. The first 5 residues are removed by prepilin peptidase. Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (strain HB27 / ATCC BAA-163 / DSM 7039) (bacteria) Thermus thermophilus (strain HB27 / ATCC BAA-163 / DSM 7039) (bacteria) |
| Molecular weight | Theoretical: 11.425754 KDa |
| Sequence | String: FTLIELAIVI VIIGILVAIA VPRFVDLTDQ ANQANVDATA AAVRSAYAIA TVQAKGIPTC DQVFANPEGG STSGSTWTSS DNSTTVSCN ASADTFTISR GGKTRTLNLT VN UniProtKB: Type IV narrow pilus major component PilA5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average electron dose: 47.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)