+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10574 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Beta-galactosidase in complex with PETG | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bgal / PETG / SUGAR BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationalkali metal ion binding / lactose catabolic process / beta-galactosidase complex / beta-galactosidase / beta-galactosidase activity / carbohydrate binding / magnesium ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Saur M / Hartshorn MJ | |||||||||
 Citation Citation |  Journal: Drug Discov Today / Year: 2020 Journal: Drug Discov Today / Year: 2020Title: Fragment-based drug discovery using cryo-EM. Authors: Michael Saur / Michael J Hartshorn / Jing Dong / Judith Reeks / Gabor Bunkoczi / Harren Jhoti / Pamela A Williams /  Abstract: Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. ...Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. Here, we review the use of cryo-EM in fragment-based drug discovery (FBDD) based on in-house method development. We demonstrate not only that cryo-EM can reveal details of the molecular interactions between fragments and a protein, but also that the current reproducibility, quality, and throughput are compatible with FBDD. We exemplify this using the test system β-galactosidase (Bgal) and the oncology target pyruvate kinase 2 (PKM2). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10574.map.gz emd_10574.map.gz | 15.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10574-v30.xml emd-10574-v30.xml emd-10574.xml emd-10574.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10574.png emd_10574.png | 172.8 KB | ||
| Filedesc metadata |  emd-10574.cif.gz emd-10574.cif.gz | 6.2 KB | ||
| Others |  emd_10574_additional.map.gz emd_10574_additional.map.gz emd_10574_additional_1.map.gz emd_10574_additional_1.map.gz emd_10574_half_map_1.map.gz emd_10574_half_map_1.map.gz emd_10574_half_map_2.map.gz emd_10574_half_map_2.map.gz | 308.8 MB 308.8 MB 261.2 MB 262.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10574 http://ftp.pdbj.org/pub/emdb/structures/EMD-10574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10574 | HTTPS FTP |
-Validation report
| Summary document |  emd_10574_validation.pdf.gz emd_10574_validation.pdf.gz | 412.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10574_full_validation.pdf.gz emd_10574_full_validation.pdf.gz | 411.4 KB | Display | |
| Data in XML |  emd_10574_validation.xml.gz emd_10574_validation.xml.gz | 496 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10574 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10574 | HTTPS FTP |
-Related structure data
| Related structure data |  6tteMC  6tshC  6tskC  6ttfC  6tthC  6ttiC  6ttqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10644 (Title: Beta-galactosidase in complex with PETG / Data size: 720.1 EMPIAR-10644 (Title: Beta-galactosidase in complex with PETG / Data size: 720.1 Data #1: Data from EPU (movies have been converted to compressed TIF) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10574.map.gz / Format: CCP4 / Size: 35.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10574.map.gz / Format: CCP4 / Size: 35.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Relion post-process unmasked map
| File | emd_10574_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-process unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
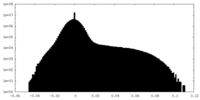
| Density Histograms |
-Additional map: Relion post-process unmasked map
| File | emd_10574_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-process unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
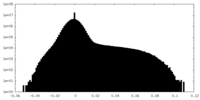
| Density Histograms |
-Half map: Relion auto-refine halfmap 1
| File | emd_10574_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion auto-refine halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Relion auto-refine halfmap 2
| File | emd_10574_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion auto-refine halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Beta-galactosidase
| Entire | Name: Beta-galactosidase |
|---|---|
| Components |
|
-Supramolecule #1: Beta-galactosidase
| Supramolecule | Name: Beta-galactosidase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 464 KDa |
-Macromolecule #1: Beta-galactosidase
| Macromolecule | Name: Beta-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: beta-galactosidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 118.395336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MLEDPVVLQR RDWENPGVTQ LNRLAAHPPF ASWRNSEEAR TDRPSQQLRS LNGEWRFAWF PAPEAVPES WLECDLPEAD TVVVPSNWQM HGYDAPIYTN VTYPITVNPP FVPTENPTGC YSLTFNVDES WLQEGQTRII F DGVNSAFH ...String: MGSSHHHHHH SSGLVPRGSH MLEDPVVLQR RDWENPGVTQ LNRLAAHPPF ASWRNSEEAR TDRPSQQLRS LNGEWRFAWF PAPEAVPES WLECDLPEAD TVVVPSNWQM HGYDAPIYTN VTYPITVNPP FVPTENPTGC YSLTFNVDES WLQEGQTRII F DGVNSAFH LWCNGRWVGY GQDSRLPSEF DLSAFLRAGE NRLAVMVLRW SDGSYLEDQD MWRMSGIFRD VSLLHKPTTQ IS DFHVATR FNDDFSRAVL EAEVQMCGEL RDYLRVTVSL WQGETQVASG TAPFGGEIID ERGGYADRVT LRLNVENPKL WSA EIPNLY RAVVELHTAD GTLIEAEACD VGFREVRIEN GLLLLNGKPL LIRGVNRHEH HPLHGQVMDE QTMVQDILLM KQNN FNAVR CSHYPNHPLW YTLCDRYGLY VVDEANIETH GMVPMNRLTD DPRWLPAMSE RVTRMVQRDR NHPSVIIWSL GNESG HGAN HDALYRWIKS VDPSRPVQYE GGGADTTATD IICPMYARVD EDQPFPAVPK WSIKKWLSLP GETRPLILCE YAHAMG NSL GGFAKYWQAF RQYPRLQGGF VWDWVDQSLI KYDENGNPWS AYGGDFGDTP NDRQFCMNGL VFADRTPHPA LTEAKHQ QQ FFQFRLSGQT IEVTSEYLFR HSDNELLHWM VALDGKPLAS GEVPLDVAPQ GKQLIELPEL PQPESAGQLW LTVRVVQP N ATAWSEAGHI SAWQQWRLAE NLSVTLPAAS HAIPHLTTSE MDFCIELGNK RWQFNRQSGF LSQMWIGDKK QLLTPLRDQ FTRAPLDNDI GVSEATRIDP NAWVERWKAA GHYQAEAALL QCTADTLADA VLITTAHAWQ HQGKTLFISR KTYRIDGSGQ MAITVDVEV ASDTPHPARI GLNCQLAQVA ERVNWLGLGP QENYPDRLTA ACFDRWDLPL SDMYTPYVFP SENGLRCGTR E LNYGPHQW RGDFQFNISR YSQQQLMETS HRHLLHAEEG TWLNIDGFHM GIGGDDSWSP SVSAEFQLSA GRYHYQLVWC QK UniProtKB: Beta-galactosidase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: 2-phenylethyl 1-thio-beta-D-galactopyranoside
| Macromolecule | Name: 2-phenylethyl 1-thio-beta-D-galactopyranoside / type: ligand / ID: 3 / Number of copies: 4 / Formula: PTQ |
|---|---|
| Molecular weight | Theoretical: 300.371 Da |
| Chemical component information |  ChemComp-PTQ: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 1224 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.17 mg/mL |
|---|---|
| Buffer | pH: 6.8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 562 / Average exposure time: 59.98 sec. / Average electron dose: 65.53 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)