+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10575 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PKM2 in complex with Compound 5 | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PKM2 / FBDD / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate kinase / pyruvate kinase activity / histone H3T11 kinase activity / programmed cell death / Pyruvate metabolism / positive regulation of cytoplasmic translation / canonical glycolysis / Glycolysis / positive regulation of sprouting angiogenesis / potassium ion binding ...pyruvate kinase / pyruvate kinase activity / histone H3T11 kinase activity / programmed cell death / Pyruvate metabolism / positive regulation of cytoplasmic translation / canonical glycolysis / Glycolysis / positive regulation of sprouting angiogenesis / potassium ion binding / rough endoplasmic reticulum / Regulation of pyruvate metabolism / glycolytic process / non-specific protein-tyrosine kinase / : / cellular response to insulin stimulus / MHC class II protein complex binding / extracellular vesicle / protein tyrosine kinase activity / secretory granule lumen / vesicle / ficolin-1-rich granule lumen / transcription coactivator activity / non-specific serine/threonine protein kinase / cilium / cadherin binding / intracellular membrane-bounded organelle / mRNA binding / Neutrophil degranulation / magnesium ion binding / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / mitochondrion / RNA binding / extracellular exosome / extracellular region / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Saur M / Hartshorn MJ | |||||||||
 Citation Citation |  Journal: Drug Discov Today / Year: 2020 Journal: Drug Discov Today / Year: 2020Title: Fragment-based drug discovery using cryo-EM. Authors: Michael Saur / Michael J Hartshorn / Jing Dong / Judith Reeks / Gabor Bunkoczi / Harren Jhoti / Pamela A Williams /  Abstract: Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. ...Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. Here, we review the use of cryo-EM in fragment-based drug discovery (FBDD) based on in-house method development. We demonstrate not only that cryo-EM can reveal details of the molecular interactions between fragments and a protein, but also that the current reproducibility, quality, and throughput are compatible with FBDD. We exemplify this using the test system β-galactosidase (Bgal) and the oncology target pyruvate kinase 2 (PKM2). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10575.map.gz emd_10575.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10575-v30.xml emd-10575-v30.xml emd-10575.xml emd-10575.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10575.png emd_10575.png | 127.3 KB | ||
| Filedesc metadata |  emd-10575.cif.gz emd-10575.cif.gz | 5.9 KB | ||
| Others |  emd_10575_additional.map.gz emd_10575_additional.map.gz emd_10575_additional_1.map.gz emd_10575_additional_1.map.gz emd_10575_half_map_1.map.gz emd_10575_half_map_1.map.gz emd_10575_half_map_2.map.gz emd_10575_half_map_2.map.gz | 163.6 MB 163.6 MB 136.8 MB 136.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10575 http://ftp.pdbj.org/pub/emdb/structures/EMD-10575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10575 | HTTPS FTP |
-Related structure data
| Related structure data |  6ttfMC  6tshC  6tskC  6tteC  6tthC  6ttiC  6ttqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10648 (Title: PKM2 in complex with Compound 5 / Data size: 773.8 EMPIAR-10648 (Title: PKM2 in complex with Compound 5 / Data size: 773.8 Data #1: Data from EPU (movies have been converted to compressed TIF) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10575.map.gz / Format: CCP4 / Size: 175 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10575.map.gz / Format: CCP4 / Size: 175 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99567 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
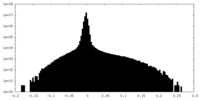
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Relion post-process unmasked map
| File | emd_10575_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-process unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
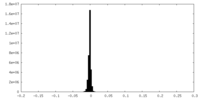
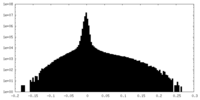
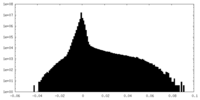
| Density Histograms |
-Additional map: Relion post-process unmasked map
| File | emd_10575_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-process unmasked map | ||||||||||||
| Projections & Slices |
| ||||||||||||
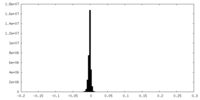
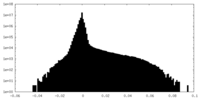
| Density Histograms |
-Half map: Relion auto-refine halfmap 1
| File | emd_10575_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion auto-refine halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
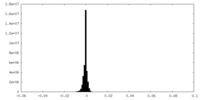
| Density Histograms |
-Half map: Relion auto-refine halfmap 2
| File | emd_10575_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion auto-refine halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
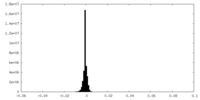
| Density Histograms |
- Sample components
Sample components
-Entire : Pyruvate Kinase M2
| Entire | Name: Pyruvate Kinase M2 |
|---|---|
| Components |
|
-Supramolecule #1: Pyruvate Kinase M2
| Supramolecule | Name: Pyruvate Kinase M2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: Pyruvate kinase PKM
| Macromolecule | Name: Pyruvate kinase PKM / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: pyruvate kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.83182 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSK PHSEAGTAFI QTQQLHAAMA DTFLEHMCRL DIDSPPITAR NTGIICTIGP ASRSVETLKE MIKSGMNVA RLNFSHGTHE YHAETIKNVR TATESFASDP ILYRPVAVAL DTKGPEIRTG LIKGSGTAEV ELKKGATLKI T LDNAYMEK ...String: MGSSHHHHHH SSGLVPRGSK PHSEAGTAFI QTQQLHAAMA DTFLEHMCRL DIDSPPITAR NTGIICTIGP ASRSVETLKE MIKSGMNVA RLNFSHGTHE YHAETIKNVR TATESFASDP ILYRPVAVAL DTKGPEIRTG LIKGSGTAEV ELKKGATLKI T LDNAYMEK CDENILWLDY KNICKVVEVG SKIYVDDGLI SLQVKQKGAD FLVTEVENGG SLGSKKGVNL PGAAVDLPAV SE KDIQDLK FGVEQDVDMV FASFIRKASD VHEVRKVLGE KGKNIKIISK IENHEGVRRF DEILEASDGI MVARGDLGIE IPA EKVFLA QKMMIGRCNR AGKPVICATQ MLESMIKKPR PTRAEGSDVA NAVLDGADCI MLSGETAKGD YPLEAVRMQH LIAR EAEAA IYHLQLFEEL RRLAPITSDP TEATAVGAVE ASFKCCSGAI IVLTKSGRSA HQVARYRPRA PIIAVTRNPQ TARQA HLYR GIFPVLCKDP VQEAWAEDVD LRVNFAMNVG KARGFFKKGD VVIVLTGWRP GSGFTNTMRV VPVP UniProtKB: Pyruvate kinase PKM |
-Macromolecule #2: 5-hydroxynaphthalene-1-sulfonamide
| Macromolecule | Name: 5-hydroxynaphthalene-1-sulfonamide / type: ligand / ID: 2 / Number of copies: 4 / Formula: LZ2 |
|---|---|
| Molecular weight | Theoretical: 223.248 Da |
| Chemical component information | 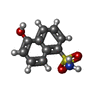 ChemComp-LZ2: |
-Macromolecule #3: 1,6-di-O-phosphono-beta-D-fructofuranose
| Macromolecule | Name: 1,6-di-O-phosphono-beta-D-fructofuranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: FBP |
|---|---|
| Molecular weight | Theoretical: 340.116 Da |
| Chemical component information | 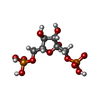 ChemComp-FBP: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 16 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.16 mg/mL |
|---|---|
| Buffer | pH: 8.2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 707 / Average exposure time: 37.06 sec. / Average electron dose: 54.85 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)