+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10389 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the RsaA N-terminal domain bound to LPS | |||||||||
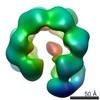
 Map data Map data | cryo-EM map of the RsaA protein N-terminal domain | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | S-layer LPS RsaA / STRUCTURAL PROTEIN | |||||||||
| Function / homology | RsaA N-terminal domain / S-layer / RTX calcium-binding nonapeptide repeat / RTX calcium-binding nonapeptide repeat (4 copies) / Serralysin-like metalloprotease, C-terminal / calcium ion binding / extracellular region / S-layer protein Function and homology information Function and homology information | |||||||||
| Biological species |  Caulobacter vibrioides CB15 (bacteria) Caulobacter vibrioides CB15 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | von Kuegelgen A / Bharat TAM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer. Authors: Andriko von Kügelgen / Haiping Tang / Gail G Hardy / Danguole Kureisaite-Ciziene / Yves V Brun / Phillip J Stansfeld / Carol V Robinson / Tanmay A M Bharat /    Abstract: Most bacterial and all archaeal cells are encapsulated by a paracrystalline, protective, and cell-shape-determining proteinaceous surface layer (S-layer). On Gram-negative bacteria, S-layers are ...Most bacterial and all archaeal cells are encapsulated by a paracrystalline, protective, and cell-shape-determining proteinaceous surface layer (S-layer). On Gram-negative bacteria, S-layers are anchored to cells via lipopolysaccharide. Here, we report an electron cryomicroscopy structure of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide. Using native mass spectrometry and molecular dynamics simulations, we deduce the length of the O-antigen on cells and show how lipopolysaccharide binding and S-layer assembly is regulated by calcium. Finally, we present a near-atomic resolution in situ structure of the complete S-layer using cellular electron cryotomography, showing S-layer arrangement at the tip of the O-antigen. A complete atomic structure of the S-layer shows the power of cellular tomography for in situ structural biology and sheds light on a very abundant class of self-assembling molecules with important roles in prokaryotic physiology with marked potential for synthetic biology and surface-display applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10389.map.gz emd_10389.map.gz | 95.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10389-v30.xml emd-10389-v30.xml emd-10389.xml emd-10389.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10389.png emd_10389.png | 19.1 KB | ||
| Filedesc metadata |  emd-10389.cif.gz emd-10389.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10389 http://ftp.pdbj.org/pub/emdb/structures/EMD-10389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10389 | HTTPS FTP |
-Related structure data
| Related structure data |  6t72MC  6z7pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10389.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10389.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of the RsaA protein N-terminal domain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of RsaA N-terminal domain bound to LPS
| Entire | Name: Structure of RsaA N-terminal domain bound to LPS |
|---|---|
| Components |
|
-Supramolecule #1: Structure of RsaA N-terminal domain bound to LPS
| Supramolecule | Name: Structure of RsaA N-terminal domain bound to LPS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Structure of RsaA N-terminal domain bound to LPS |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides CB15 (bacteria) / Strain: YB1001 Caulobacter vibrioides CB15 (bacteria) / Strain: YB1001 |
| Molecular weight | Theoretical: 577 KDa |
-Macromolecule #1: S-layer protein
| Macromolecule | Name: S-layer protein / type: protein_or_peptide / ID: 1 / Details: LPS O-antigen bound to protein / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides CB15 (bacteria) Caulobacter vibrioides CB15 (bacteria) |
| Molecular weight | Theoretical: 25.820354 KDa |
| Recombinant expression | Organism:  Caulobacter vibrioides CB15 (bacteria) Caulobacter vibrioides CB15 (bacteria) |
| Sequence | String: AYTTAQLVTA YTNANLGKAP DAATTLTLDA YATQTQTGGL SDAAALTNTL KLVNSTTAVA IQTYQFFTGV APSAAGLDFL VDSTTNTND LNDAYYSKFA QENRFINFSI NLATGAGAGA TAFAAAYTGV SYAQTVATAY DKIIGNAVAT AAGVDVAAAV A FLSRQANI ...String: AYTTAQLVTA YTNANLGKAP DAATTLTLDA YATQTQTGGL SDAAALTNTL KLVNSTTAVA IQTYQFFTGV APSAAGLDFL VDSTTNTND LNDAYYSKFA QENRFINFSI NLATGAGAGA TAFAAAYTGV SYAQTVATAY DKIIGNAVAT AAGVDVAAAV A FLSRQANI DYLTAFVRAN TPFTAAADID LAVKAALIGT ILNAATVSGI GGYATATAAM INDLSDGALS TDNAAGVNLF TA YPSSGVS GSENLYFQ UniProtKB: S-layer protein |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Buffer solutions were prepared fresh from sterile filtered concentrated stocksolutions. Solutions were filtered through a 0.22 um filter to avoid microbial contamination and degassed using a ...Details: Buffer solutions were prepared fresh from sterile filtered concentrated stocksolutions. Solutions were filtered through a 0.22 um filter to avoid microbial contamination and degassed using a vacuum fold pump. The pH of the HEPES stock solution was adjusted with sodium hydroxide at 4 deg C. | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Details: 20 seconds, 15 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Vitrobot options: Blot time 3 seconds, Blot force -13,1, Wait time 10 seconds, Drain time 0.5 seconds,. | |||||||||||||||
| Details | RsaA N-terminal domain with LPS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Details | EPU software |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-20 / Number grids imaged: 1 / Number real images: 2422 / Average exposure time: 8.0 sec. / Average electron dose: 43.0 e/Å2 Details: Images were collected in movie-mode and subjected to 8 seconds of exposure where a total dose of 43 e-/A2 was applied, and 20 frames were recorded per movie. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: -4.0 µm / Calibrated defocus min: -1.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -4.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | The carbon backbone of the RsaA protein was manually traced through a single subunit of the cryo-EM density using Coot (Emsley et al., 2010). Initially, side chains were assigned in regions with density corresponding to characteristic aromatic residues allowing us to deduce the register of the amino acid sequence in the map. Side chains for residues 2-243 of RsaA were thus assigned unambiguously and the structure was refined and manually rebuilt using Refmac5 (Murshudov et al., 2011) inside the CCP-EM (Burnley et al., 2017) software suite and Coot. |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: BACKBONE TRACE / Overall B value: 85.819 / Target criteria: Best fit |
| Output model |  PDB-6t72: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















