[English] 日本語
 Yorodumi
Yorodumi- EMDB-10243: Structure of the RagAB peptide importer in the 'open-open' state -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10243 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the RagAB peptide importer in the 'open-open' state | ||||||||||||||||||||||||
 Map data Map data | Sharpened map filtered by local resolution in RELION | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Beta-barrel / OMP / TonB-dependent / transporter / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |  Porphyromonas gingivalis W83 (bacteria) Porphyromonas gingivalis W83 (bacteria) | ||||||||||||||||||||||||
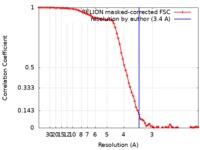
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||||||||
 Authors Authors | White JBR / Ranson NA | ||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Poland, 7 items Poland, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2020 Journal: Nat Microbiol / Year: 2020Title: Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Authors: Mariusz Madej / Joshua B R White / Zuzanna Nowakowska / Shaun Rawson / Carsten Scavenius / Jan J Enghild / Grzegorz P Bereta / Karunakar Pothula / Ulrich Kleinekathoefer / Arnaud Baslé / ...Authors: Mariusz Madej / Joshua B R White / Zuzanna Nowakowska / Shaun Rawson / Carsten Scavenius / Jan J Enghild / Grzegorz P Bereta / Karunakar Pothula / Ulrich Kleinekathoefer / Arnaud Baslé / Neil A Ranson / Jan Potempa / Bert van den Berg /      Abstract: Porphyromonas gingivalis, an asaccharolytic member of the Bacteroidetes, is a keystone pathogen in human periodontitis that may also contribute to the development of other chronic inflammatory ...Porphyromonas gingivalis, an asaccharolytic member of the Bacteroidetes, is a keystone pathogen in human periodontitis that may also contribute to the development of other chronic inflammatory diseases. P. gingivalis utilizes protease-generated peptides derived from extracellular proteins for growth, but how these peptides enter the cell is not clear. Here, we identify RagAB as the outer-membrane importer for these peptides. X-ray crystal structures show that the transporter forms a dimeric RagAB complex, with the RagB substrate-binding surface-anchored lipoprotein forming a closed lid on the RagA TonB-dependent transporter. Cryo-electron microscopy structures reveal the opening of the RagB lid and thus provide direct evidence for a 'pedal bin' mechanism of nutrient uptake. Together with mutagenesis, peptide-binding studies and RagAB peptidomics, our work identifies RagAB as a dynamic, selective outer-membrane oligopeptide-acquisition machine that is essential for the efficient utilization of proteinaceous nutrients by P. gingivalis. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10243.map.gz emd_10243.map.gz | 24.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10243-v30.xml emd-10243-v30.xml emd-10243.xml emd-10243.xml | 30.3 KB 30.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10243_fsc.xml emd_10243_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10243.png emd_10243.png | 61.7 KB | ||
| Masks |  emd_10243_msk_1.map emd_10243_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10243.cif.gz emd-10243.cif.gz | 8 KB | ||
| Others |  emd_10243_additional_1.map.gz emd_10243_additional_1.map.gz emd_10243_additional_2.map.gz emd_10243_additional_2.map.gz emd_10243_additional_3.map.gz emd_10243_additional_3.map.gz | 14.2 MB 14.2 MB 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10243 http://ftp.pdbj.org/pub/emdb/structures/EMD-10243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10243 | HTTPS FTP |
-Related structure data
| Related structure data |  6smlMC  6sliC  6sljC  6slnC  6sm3C  6smqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10543 (Title: Cryo electron microscopy of RagAB from P. gingivalis solubilised in DDM EMPIAR-10543 (Title: Cryo electron microscopy of RagAB from P. gingivalis solubilised in DDMData size: 9.0 TB Data #1: Raw micrograph movies of the RagAB complex solubilised in DDM [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10243.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10243.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map filtered by local resolution in RELION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10243_msk_1.map emd_10243_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
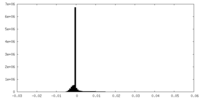
| Density Histograms |
-Additional map: Half map 1 from final iteration of 3D auto-refinement in RELION
| File | emd_10243_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from final iteration of 3D auto-refinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
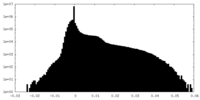
| Density Histograms |
-Additional map: Half map 2 from final iteration of 3D auto-refinement in RELION
| File | emd_10243_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from final iteration of 3D auto-refinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
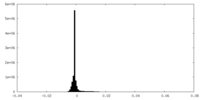
| Density Histograms |
-Additional map: Unsharpened map from 3D auto-refinement in RELION
| File | emd_10243_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from 3D auto-refinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
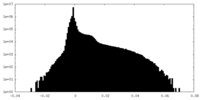
| Density Histograms |
- Sample components
Sample components
-Entire : RagAB with putative peptide substrate
| Entire | Name: RagAB with putative peptide substrate |
|---|---|
| Components |
|
-Supramolecule #1: RagAB with putative peptide substrate
| Supramolecule | Name: RagAB with putative peptide substrate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Putative peptide substrate co-purified with the complex |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB |
-Macromolecule #1: Lipoprotein RagB
| Macromolecule | Name: Lipoprotein RagB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB |
| Molecular weight | Theoretical: 54.430863 KDa |
| Sequence | String: CELDRDPEGK DFQQPYTSFV QTKQNRDGLY ALLRNTENPR MHFYQELQSD MYCTTITDGN SLAPFVNWDL GILNDHGRAD EDEVSGIAG YYFVYNRLNQ QANAFVNNTE AALQNQVYKN STEIANAKSF LAEGKVLQAL AIWRLMDRFS FHESVTEVNS G AKDLGVIL ...String: CELDRDPEGK DFQQPYTSFV QTKQNRDGLY ALLRNTENPR MHFYQELQSD MYCTTITDGN SLAPFVNWDL GILNDHGRAD EDEVSGIAG YYFVYNRLNQ QANAFVNNTE AALQNQVYKN STEIANAKSF LAEGKVLQAL AIWRLMDRFS FHESVTEVNS G AKDLGVIL LKEYNPGYIG PRATKAQCYD YILSRLSEAI EVLPENRESV LYVSRDYAYA LRARIYLALG EYGKAAADAK MV VDKYPLI GAADASEFEN IYRSDANNPE IIFRGFASAT LGSFTATTLN GAAPAGKDIK YNPSAVPFQW VVDLYENEDF RKS VYIAKV VKKDKGYLVN KFLEDKAYRD VQDKPNLKVG ARYFSVAEVY LILVESALQT GDTPTAEKYL KALSKARGAE VSVV NMEAL QAERTRELIG EGSRLRDMVR WSIPNNHDAF ETQPGLEGFA NTTPLKAQAP VGFYAYTWEF PQRDRQTNPQ LIKNW PI UniProtKB: Lipoprotein RagB |
-Macromolecule #2: RagA protein
| Macromolecule | Name: RagA protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB |
| Molecular weight | Theoretical: 101.582398 KDa |
| Sequence | String: QVVVLGYGTG QKLSTVSGSV AKVSSEKLAE KPVANIMDAL QGQVAGMQVM TTSGDPTAVA SVEIHGTGSL GASSAPLYIV DGMQTSLDV VATMNPNDFE SMSVLKDASA TSIYGARAAN GVVFIQTKKG KMSERGRITF NASYGISQIL NTKPLDNMMT G DELLDFQV ...String: QVVVLGYGTG QKLSTVSGSV AKVSSEKLAE KPVANIMDAL QGQVAGMQVM TTSGDPTAVA SVEIHGTGSL GASSAPLYIV DGMQTSLDV VATMNPNDFE SMSVLKDASA TSIYGARAAN GVVFIQTKKG KMSERGRITF NASYGISQIL NTKPLDNMMT G DELLDFQV KAGFWGNNQT VQKVKDMILA GAEDLYGNYD SLKDEYGKTL FPVDFNHDAD WLKALFKTAP TSQGDISFSG GS QGTSYYA SIGYFDQEGM AREPANFKRY SGRLNFESRI NEWLKVGANL SGAIANRRSA DYFGKYYMGS GTFGVLTMPR YYN PFDVNG DLADVYYMYG ATRPSMTEPY FAKMRPFSSE SHQANVNGFA QITPIKGLTL KAQAGVDITN TRTSSKRMPN NPYD STPLG ERRERAYRDV SKSFTNTAEY KFSIDEKHDL TALMGHEYIE YEGDVIGASS KGFESDKLML LSQGKTGNSL SLPEH RVAE YAYLSFFSRF NYGFDKWMYI DFSVRNDQSS RFGSNNRSAW FYSVGGMFDI YNKFIQESNW LSDLRLKMSY GTTGNS EIG NYNHQALVTV NNYTEDAMGL SISTAGNPDL SWEKQSQFNF GLAAGAFNNR LSAEVDFYVR TTNDMLIDVP MPYISGF FS QYQNVGSMKN TGVDLSLKGT IYQNKDWNVY ASANFNYNRQ EITKLFFGLN KYMLPNTGTI WEIGYPNSFY MAEYAGID K KTGKQLWYVP GQVDADGNKV TTSQYSADLE TRIDKSVTPP ITGGFSLGAS WKGLSLDADF AYIVGKWMIN NDRYFTENG GGLMQLNKDK MLLNAWTEDN KETDVPKLGQ SPQFDTHLLE NASFLRLKNL KLTYVLPNSL FAGQNVIGGA RVYLMARNLL TVTKYKGFD PEAGGNVGKN QYPNSKQYVA GIQLSF UniProtKB: RagA protein |
-Macromolecule #3: GLY-THR-GLY-GLY-SER-THR-GLY-THR-THR-SER-ALA-GLY
| Macromolecule | Name: GLY-THR-GLY-GLY-SER-THR-GLY-THR-THR-SER-ALA-GLY / type: protein_or_peptide / ID: 3 / Details: Putative peptide substrate / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB Porphyromonas gingivalis W83 (bacteria) / Strain: KRAB |
| Molecular weight | Theoretical: 952.923 Da |
| Sequence | String: GTGGSTGTTS AG |
-Macromolecule #4: (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OX...
| Macromolecule | Name: (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE type: ligand / ID: 4 / Number of copies: 1 / Formula: 5PL |
|---|---|
| Molecular weight | Theoretical: 1.233719 KDa |
| Chemical component information |  ChemComp-5PL: |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.75 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279.15 K / Instrument: FEI VITROBOT MARK IV / Details: 6 second blot time, blot force 6. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 77.88 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)