+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-10213 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
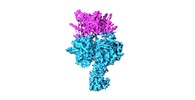
| タイトル | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to RBM39 and Indisulam | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | DDB1 / ONCOPROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報RS domain binding / regulation of natural killer cell activation / positive regulation by virus of viral protein levels in host cell / U1 snRNP binding / regulation of mRNA splicing, via spliceosome / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition ...RS domain binding / regulation of natural killer cell activation / positive regulation by virus of viral protein levels in host cell / U1 snRNP binding / regulation of mRNA splicing, via spliceosome / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / biological process involved in interaction with symbiont / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / small molecule binding / negative regulation of reproductive process / negative regulation of developmental process / cullin family protein binding / viral release from host cell / immune system process / ectopic germ cell programmed cell death / RNA processing / positive regulation of viral genome replication / proteasomal protein catabolic process / positive regulation of gluconeogenesis / mRNA Splicing - Major Pathway / RNA splicing / nucleotide-excision repair / Recognition of DNA damage by PCNA-containing replication complex / regulation of circadian rhythm / DNA Damage Recognition in GG-NER / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / centriolar satellite / Formation of Incision Complex in GG-NER / Wnt signaling pathway / mRNA processing / protein polyubiquitination / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / positive regulation of protein catabolic process / cellular response to UV / rhythmic process / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / site of double-strand break / Neddylation / microtubule cytoskeleton / ubiquitin-dependent protein catabolic process / protein-macromolecule adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / damaged DNA binding / chromosome, telomeric region / nuclear speck / protein ubiquitination / DNA repair / apoptotic process / DNA damage response / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / protein-containing complex / extracellular space / DNA binding / RNA binding / extracellular exosome / nucleoplasm / metal ion binding / nucleus / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
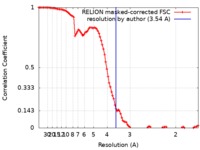
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.54 Å | |||||||||
 データ登録者 データ登録者 | Srinivas H | |||||||||
 引用 引用 |  ジャーナル: Nat Chem Biol / 年: 2020 ジャーナル: Nat Chem Biol / 年: 2020タイトル: Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. 著者: Dirksen E Bussiere / Lili Xie / Honnappa Srinivas / Wei Shu / Ashley Burke / Celine Be / Junping Zhao / Adarsh Godbole / Dan King / Rajeshri G Karki / Viktor Hornak / Fangmin Xu / Jennifer ...著者: Dirksen E Bussiere / Lili Xie / Honnappa Srinivas / Wei Shu / Ashley Burke / Celine Be / Junping Zhao / Adarsh Godbole / Dan King / Rajeshri G Karki / Viktor Hornak / Fangmin Xu / Jennifer Cobb / Nathalie Carte / Andreas O Frank / Alexandra Frommlet / Patrick Graff / Mark Knapp / Aleem Fazal / Barun Okram / Songchun Jiang / Pierre-Yves Michellys / Rohan Beckwith / Hans Voshol / Christian Wiesmann / Jonathan M Solomon / Joshiawa Paulk /   要旨: The anticancer agent indisulam inhibits cell proliferation by causing degradation of RBM39, an essential mRNA splicing factor. Indisulam promotes an interaction between RBM39 and the DCAF15 E3 ligase ...The anticancer agent indisulam inhibits cell proliferation by causing degradation of RBM39, an essential mRNA splicing factor. Indisulam promotes an interaction between RBM39 and the DCAF15 E3 ligase substrate receptor, leading to RBM39 ubiquitination and proteasome-mediated degradation. To delineate the precise mechanism by which indisulam mediates the DCAF15-RBM39 interaction, we solved the DCAF15-DDB1-DDA1-indisulam-RBM39(RRM2) complex structure to a resolution of 2.3 Å. DCAF15 has a distinct topology that embraces the RBM39(RRM2) domain largely via non-polar interactions, and indisulam binds between DCAF15 and RBM39(RRM2), coordinating additional interactions between the two proteins. Studies with RBM39 point mutants and indisulam analogs validated the structural model and defined the RBM39 α-helical degron motif. The degron is found only in RBM23 and RBM39, and only these proteins were detectably downregulated in indisulam-treated HCT116 cells. This work further explains how indisulam induces RBM39 degradation and defines the challenge of harnessing DCAF15 to degrade additional targets. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_10213.map.gz emd_10213.map.gz | 5.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-10213-v30.xml emd-10213-v30.xml emd-10213.xml emd-10213.xml | 16.1 KB 16.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_10213_fsc.xml emd_10213_fsc.xml | 10 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_10213.png emd_10213.png | 115.3 KB | ||
| Filedesc metadata |  emd-10213.cif.gz emd-10213.cif.gz | 6.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10213 http://ftp.pdbj.org/pub/emdb/structures/EMD-10213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10213 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_10213_validation.pdf.gz emd_10213_validation.pdf.gz | 381.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_10213_full_validation.pdf.gz emd_10213_full_validation.pdf.gz | 380.7 KB | 表示 | |
| XML形式データ |  emd_10213_validation.xml.gz emd_10213_validation.xml.gz | 11.3 KB | 表示 | |
| CIF形式データ |  emd_10213_validation.cif.gz emd_10213_validation.cif.gz | 15.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10213 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10213 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10213 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10213 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_10213.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_10213.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : DDB1 complex
| 全体 | 名称: DDB1 complex |
|---|---|
| 要素 |
|
-超分子 #1: DDB1 complex
| 超分子 | 名称: DDB1 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#4 |
|---|---|
| 分子量 | 理論値: 210 KDa |
-超分子 #2: DDB1-DDA1-DCAF15-RBM39 complex
| 超分子 | 名称: DDB1-DDA1-DCAF15-RBM39 complex / タイプ: complex / ID: 2 / 親要素: 1 / 含まれる分子: #1-#4 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-超分子 #3: RBM39
| 超分子 | 名称: RBM39 / タイプ: complex / ID: 3 / 親要素: 1 / 含まれる分子: #3 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: DDB1- and CUL4-associated factor 15
| 分子 | 名称: DDB1- and CUL4-associated factor 15 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 66.563297 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GPAPSSKSER NSGAGSGGGG PGGAGGKRAA GRRREHVLKQ LERVKISGQL SPRLFRKLPP RVCVSLKNIV DEDFLYAGHI FLGFSKCGR YVLSYTSSSG DDDFSFYIYH LYWWEFNVHS KLKLVRQVRL FQDEEIYSDL YLTVCEWPSD ASKVIVFGFN T RSANGMLM ...文字列: GPAPSSKSER NSGAGSGGGG PGGAGGKRAA GRRREHVLKQ LERVKISGQL SPRLFRKLPP RVCVSLKNIV DEDFLYAGHI FLGFSKCGR YVLSYTSSSG DDDFSFYIYH LYWWEFNVHS KLKLVRQVRL FQDEEIYSDL YLTVCEWPSD ASKVIVFGFN T RSANGMLM NMMMMSDENH RDIYVSTVAV PPPGRCAACQ DASRAHPGDP NAQCLRHGFM LHTKYQVVYP FPTFQPAFQL KK DQVVLLN TSYSLVACAV SVHSAGDRSF CQILYDHSTC PLAPASPPEP QSPELPPALP SFCPEAAPAR SSGSPEPSPA IAK AKEFVA DIFRRAKEAK GGVPEEARPA LCPGPSGSRC RAHSEPLALC GETAPRDSPP ASEAPASEPG YVNYTKLYYV LESG EGTEP EDELEDDKIS LPFVVTDLRG RNLRPMRERT AVQGQYLTVE QLTLDFEYVI NEVIRHDATW GHQFCSFSDY DIVIL EVCP ETNQVLINIG LLLLAFPSPT EEGQLRPKTY HTSLKVAWDL NTGIFETVSV GDLTEVKGQT SGSVWSSYRK SCVDMV MKW LVPESSGRYV NRMTNEALHK GCSLKVLADS ERYTWIVL UniProtKB: DDB1- and CUL4-associated factor 15 |
-分子 #2: DNA damage-binding protein 1
| 分子 | 名称: DNA damage-binding protein 1 / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 127.097469 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK ...文字列: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK FLYGCQAPTI CFVYQDPQGR HVKTYEVSLR EKEFNKGPWK QENVEAEASM VIAVPEPFGG AIIIGQESIT YH NGDKYLA IAPPIIKQST IVCHNRVDPN GSRYLLGDME GRLFMLLLEK EEQMDGTVTL KDLRVELLGE TSIAECLTYL DNG VVFVGS RLGDSQLVKL NVDSNEQGSY VVAMETFTNL GPIVDMCVVD LERQGQGQLV TCSGAFKEGS LRIIRNGIGI HEHA SIDLP GIKGLWPLRS DPNRETDDTL VLSFVGQTRV LMLNGEEVEE TELMGFVDDQ QTFFCGNVAH QQLIQITSAS VRLVS QEPK ALVSEWKEPQ AKNISVASCN SSQVVVAVGR ALYYLQIHPQ ELRQISHTEM EHEVACLDIT PLGDSNGLSP LCAIGL WTD ISARILKLPS FELLHKEMLG GEIIPRSILM TTFESSHYLL CALGDGALFY FGLNIETGLL SDRKKVTLGT QPTVLRT FR SLSTTNVFAC SDRPTVIYSS NHKLVFSNVN LKEVNYMCPL NSDGYPDSLA LANNSTLTIG TIDEIQKLHI RTVPLYES P RKICYQEVSQ CFGVLSSRIE VQDTSGGTTA LRPSASTQAL SSSVSSSKLF SSSTAPHETS FGEEVEVHNL LIIDQHTFE VLHAHQFLQN EYALSLVSCK LGKDPNTYFI VGTAMVYPEE AEPKQGRIVV FQYSDGKLQT VAEKEVKGAV YSMVEFNGKL LASINSTVR LYEWTTEKEL RTECNHYNNI MALYLKTKGD FILVGDLMRS VLLLAYKPME GNFEEIARDF NPNWMSAVEI L DDDNFLGA ENAFNLFVCQ KDSAATTDEE RQHLQEVGLF HLGEFVNVFC HGSLVMQNLG ETSTPTQGSV LFGTVNGMIG LV TSLSESW YNLLLDMQNR LNKVIKSVGK IEHSFWRSFH TERKTEPATG FIDGDLIESF LDISRPKMQE VVANLQYDDG SGM KREATA DDLIKVVEEL TRIH UniProtKB: DNA damage-binding protein 1 |
-分子 #3: RNA binding protein 39
| 分子 | 名称: RNA binding protein 39 / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 9.085409 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GPMRLYVGSL HFNITEDMLR GIFEPFGRIE SIQLMMDSET GRSKGYGFIT FSDSECAKKA LEQLNGFELA GRPMKVGHVT E UniProtKB: Uncharacterized protein DKFZp781I1140 |
-分子 #4: DET1- and DDB1-associated protein 1
| 分子 | 名称: DET1- and DDB1-associated protein 1 / タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 11.724102 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: ADFLKGLPVY NKSNFSRFHA DSVCKASNRR PSVYLPTREY PSEQIIVTEK TNILLRYLHQ QWDKKNAAKK RDQEQVELEG ESSAPPRKV ARTDSPDMHE DT UniProtKB: DET1- and DDB1-associated protein 1 |
-分子 #5: N~1~-(3-chloro-1H-indol-7-yl)benzene-1,4-disulfonamide
| 分子 | 名称: N~1~-(3-chloro-1H-indol-7-yl)benzene-1,4-disulfonamide タイプ: ligand / ID: 5 / コピー数: 1 / 式: EF6 |
|---|---|
| 分子量 | 理論値: 385.846 Da |
| Chemical component information | 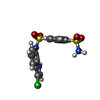 ChemComp-EF6: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 QUANTUM (4k x 4k) 検出モード: COUNTING / 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)