+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10177 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Body domain of the mt-SSU assemblosome from Trypanosoma brucei | |||||||||||||||
 Map data Map data | Body domain of the mt-SSU assemblosome of Trypanosoma brucei brucei | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | mitochondrial ribosome / assembly intermediate / translation / RIBOSOME | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmodulation of formation of structure involved in a symbiotic process / rRNA (cytosine-N4-)-methyltransferase activity / ATP-activated inward rectifier potassium channel activity / quorum sensing / kinetoplast / response to arsenic-containing substance / ciliary plasm / rRNA base methylation / mitochondrial small ribosomal subunit / acyl binding ...modulation of formation of structure involved in a symbiotic process / rRNA (cytosine-N4-)-methyltransferase activity / ATP-activated inward rectifier potassium channel activity / quorum sensing / kinetoplast / response to arsenic-containing substance / ciliary plasm / rRNA base methylation / mitochondrial small ribosomal subunit / acyl binding / acyl carrier activity / iron-sulfur cluster binding / voltage-gated potassium channel activity / axoneme / voltage-gated potassium channel complex / Transferases; Transferring one-carbon groups; Methyltransferases / methyltransferase activity / protein homooligomerization / potassium ion transport / fatty acid biosynthetic process / structural constituent of ribosome / translation / mitochondrial matrix / mRNA binding / mitochondrion / metal ion binding / membrane / cytosol / cytoplasm Similarity search - Function T-cell activation inhibitor, mitochondrial / Domain of unknown function DUF4460 / Domain of unknown function (DUF4460) / Probable Zinc-ribbon domain / Probable Treble clef zinc finger domain / Ribosomal protein Rsm22-like / : / Mitochondrial small ribosomal subunit Rsm22 / Ribosomal RNA small subunit methyltransferase H / S-adenosyl-L-methionine-dependent methyltransferase, MraW, recognition domain superfamily ...T-cell activation inhibitor, mitochondrial / Domain of unknown function DUF4460 / Domain of unknown function (DUF4460) / Probable Zinc-ribbon domain / Probable Treble clef zinc finger domain / Ribosomal protein Rsm22-like / : / Mitochondrial small ribosomal subunit Rsm22 / Ribosomal RNA small subunit methyltransferase H / S-adenosyl-L-methionine-dependent methyltransferase, MraW, recognition domain superfamily / MraW methylase family / Protein Fyv4 / Small ribosomal subunit protein mS41 SAM domain / IGR protein motif / IGR / Phosphotyrosine protein phosphatase I / Phosphotyrosine protein phosphatase I superfamily / Low molecular weight phosphotyrosine protein phosphatase / Low molecular weight phosphatase family / Pentatricopeptide repeat domain / Pentatricopeptide (PPR) repeat profile. / Pentatricopeptide repeat / Potassium channel tetramerisation-type BTB domain / BTB/POZ domain / Acyl carrier protein (ACP) / Phosphopantetheine attachment site / SKP1/BTB/POZ domain superfamily / Phosphopantetheine attachment site. / Phosphopantetheine attachment site / ACP-like superfamily / Carrier protein (CP) domain profile. / Phosphopantetheine binding ACP domain / Translation elongation factor EF1B/ribosomal protein S6 / Tetratricopeptide-like helical domain superfamily / Ribosomal protein S11 superfamily / S15/NS1, RNA-binding / Thioredoxin-like superfamily / S-adenosyl-L-methionine-dependent methyltransferase superfamily Similarity search - Domain/homology Uncharacterized protein / Uncharacterized protein / Methyltransferase / Uncharacterized protein / Potassium voltage-gated channel, putative / Uncharacterized protein / Small ribosomal subunit protein mS41 / Uncharacterized protein / Uncharacterized protein / Uncharacterized protein ...Uncharacterized protein / Uncharacterized protein / Methyltransferase / Uncharacterized protein / Potassium voltage-gated channel, putative / Uncharacterized protein / Small ribosomal subunit protein mS41 / Uncharacterized protein / Uncharacterized protein / Uncharacterized protein / Uncharacterized protein / Target SNARE / Phosphotyrosine protein phosphatase I domain-containing protein / DUF4460 domain-containing protein / Uncharacterized protein / Uncharacterized protein / Pentacotripeptide-repeat region of PRORP domain-containing protein / Uncharacterized protein / Glutaredoxin domain-containing protein / S-adenosyl-methyltransferase mraW-like protein, putative / Acyl carrier protein / Pentacotripeptide-repeat region of PRORP domain-containing protein / Treble clef zinc finger domain-containing protein / Succinate dehydrogenase assembly factor 3, mitochondrial / Uncharacterized protein Similarity search - Component | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||
 Authors Authors | Saurer M / Ramrath DJF | |||||||||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Authors: Martin Saurer / David J F Ramrath / Moritz Niemann / Salvatore Calderaro / Céline Prange / Simone Mattei / Alain Scaiola / Alexander Leitner / Philipp Bieri / Elke K Horn / Marc Leibundgut ...Authors: Martin Saurer / David J F Ramrath / Moritz Niemann / Salvatore Calderaro / Céline Prange / Simone Mattei / Alain Scaiola / Alexander Leitner / Philipp Bieri / Elke K Horn / Marc Leibundgut / Daniel Boehringer / André Schneider / Nenad Ban /  Abstract: Mitochondrial ribosomes (mitoribosomes) are large ribonucleoprotein complexes that synthesize proteins encoded by the mitochondrial genome. An extensive cellular machinery responsible for ribosome ...Mitochondrial ribosomes (mitoribosomes) are large ribonucleoprotein complexes that synthesize proteins encoded by the mitochondrial genome. An extensive cellular machinery responsible for ribosome assembly has been described only for eukaryotic cytosolic ribosomes. Here we report that the assembly of the small mitoribosomal subunit in involves a large number of factors and proceeds through the formation of assembly intermediates, which we analyzed by using cryo-electron microscopy. One of them is a 4-megadalton complex, referred to as the small subunit assemblosome, in which we identified 34 factors that interact with immature ribosomal RNA (rRNA) and recognize its functionally important regions. The assembly proceeds through large-scale conformational changes in rRNA coupled with successive incorporation of mitoribosomal proteins, providing an example for the complexity of the ribosomal assembly process in mitochondria. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10177.map.gz emd_10177.map.gz | 17.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10177-v30.xml emd-10177-v30.xml emd-10177.xml emd-10177.xml | 96.7 KB 96.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10177.png emd_10177.png | 59.7 KB | ||
| Filedesc metadata |  emd-10177.cif.gz emd-10177.cif.gz | 21 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10177 http://ftp.pdbj.org/pub/emdb/structures/EMD-10177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10177 | HTTPS FTP |
-Related structure data
| Related structure data |  6sgaMC  6sg9C  6sgbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10177.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10177.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body domain of the mt-SSU assemblosome of Trypanosoma brucei brucei | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
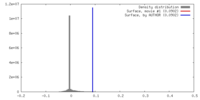
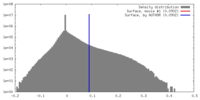
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Body domain of the mt-SSU assemblosome
| Entire | Name: Body domain of the mt-SSU assemblosome |
|---|---|
| Components |
|
+Supramolecule #1: Body domain of the mt-SSU assemblosome
| Supramolecule | Name: Body domain of the mt-SSU assemblosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#62 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: uS5m
| Macromolecule | Name: uS5m / type: protein_or_peptide / ID: 1 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.147355 KDa |
| Sequence | String: MFRLTAASQK NLNSWYTKGT MRGGVPRIYY AWMRPGSFTR RRFEKMRNPF VDLETGTSLY FRDTRDSAEA IAHAADSKGI KGMDNAIDL YNEYRIVPDL YPEGFQWKHK LNTEYNQWRS NTWLTPDLIP KEHRGRFLCN FQLNIVAYDM RVVKFSPKDH R QWIYCVLY ...String: MFRLTAASQK NLNSWYTKGT MRGGVPRIYY AWMRPGSFTR RRFEKMRNPF VDLETGTSLY FRDTRDSAEA IAHAADSKGI KGMDNAIDL YNEYRIVPDL YPEGFQWKHK LNTEYNQWRS NTWLTPDLIP KEHRGRFLCN FQLNIVAYDM RVVKFSPKDH R QWIYCVLY VGSGKGIAGW GRAVAPSTQE AKKEAIREAF SNIIAVDLEQ EGPMYPVRVN ADGVRVLLYP ARRIVANFRV AD ILCAFGF QHAGCRINLK ATNNPKSPTH TVEGVFEAVK ALRSVSEIAA SRGKVPHSLI YNIYPYLEEI RRRKGMMAMH PPG KDGLLM PDRVVDNRLP DHLKRGYYDD VYWKDFFAGS DEHLNEPRMG LRGDEMRRRL EEAQTSPAPT TAKDTRRRTL EDVL KRLGK TTRDLGSIPI VNPRVDVGLP THIKRNYQLH |
+Macromolecule #2: bS6m
| Macromolecule | Name: bS6m / type: protein_or_peptide / ID: 2 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.861543 KDa |
| Sequence | String: MVFYSFVLVM KPRQRRFTSQ ALREIGVAVY SNGGLIRSIT NEGIMRPYSR FRDADNTPLT YARYIILQLD MGEEEMGKVD KIIREHQDV LMALKLNNLE RPVGIRSGNK ELQAAYFPLD TFTRLEEEIN WSPQTSADIY TQLEMNWKEF SRTRWSSFLR N UniProtKB: Uncharacterized protein |
+Macromolecule #3: uS8m
| Macromolecule | Name: uS8m / type: protein_or_peptide / ID: 3 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.766459 KDa |
| Sequence | String: MLRQSLLSLA KVYGAVPPLG AAQGHRLLHG KREREGSLFA VANDVKRDER LLRQQLNALL EEERMPMQAR LQRNEAVSSH EAVPQTPLV DLPGVERRRD LPADPITRLF FQHKGDHALY YGTYDKPSVQ DDDRVQIEKR VPRRREENLY TPIYDFCHRI R EATEQRKR ...String: MLRQSLLSLA KVYGAVPPLG AAQGHRLLHG KREREGSLFA VANDVKRDER LLRQQLNALL EEERMPMQAR LQRNEAVSSH EAVPQTPLV DLPGVERRRD LPADPITRLF FQHKGDHALY YGTYDKPSVQ DDDRVQIEKR VPRRREENLY TPIYDFCHRI R EATEQRKR FVVVPSTIET RGCARVMHDH GLVAGFRDFH NDRAFAVELK YFQGDSTINV IEPCSYDGRT EFEWSPKMMR RL LNTHGIH NRLVVYICRT ADNRIIDHIH AVKENIGGRG LMMVH |
+Macromolecule #4: uS11m
| Macromolecule | Name: uS11m / type: protein_or_peptide / ID: 4 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.953965 KDa |
| Sequence | String: MMRKSVTFLR RGKPRPRAGM FPDKYRRVPM LLKPQQGGQQ YFNHFLIRST NDRLTQQDVD NASGQAHSFI SPQLPQMDWR NMSARSSEE SIREEMRQLA ENDVMQHQRV FNERMWYEKE EEHRMKARSC PEETSEHAIS NDGAVPPPRV LGGDYFKTRF G YSLVKNSE ...String: MMRKSVTFLR RGKPRPRAGM FPDKYRRVPM LLKPQQGGQQ YFNHFLIRST NDRLTQQDVD NASGQAHSFI SPQLPQMDWR NMSARSSEE SIREEMRQLA ENDVMQHQRV FNERMWYEKE EEHRMKARSC PEETSEHAIS NDGAVPPPRV LGGDYFKTRF G YSLVKNSE MTQGPVDYSQ LDMWGEMPRY TSDMVFLYLV SRRRNTYAVA YTYEGKRILN TYTAGNRGLK GGDRGFRSEG ST DNGHQVT SMYLNDLLPK LREMRASEGR PMGRGEKVEL VVRVMGFYNG RQGAVRAVQD RANEFHVRYF EDITPFPLNG PKM PRGVFK UniProtKB: Uncharacterized protein |
+Macromolecule #5: uS15m
| Macromolecule | Name: uS15m / type: protein_or_peptide / ID: 5 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.401191 KDa |
| Sequence | String: MFVRSFCFLR KYGSKSKTPD LKTSFTKTTN FRKAEKISIT TTDGEIVKGR GKIRRRTHSA LKEAIGAMEH PAIWLWYPWR MNPEPPTPH MPQRRALKNV HGAVFNDLTP VQKKRQEQML YGVNIPETRQ MKFEEQHPLL AGALRKLEGQ PKGFPFWYRK Y PTRRHAYE ...String: MFVRSFCFLR KYGSKSKTPD LKTSFTKTTN FRKAEKISIT TTDGEIVKGR GKIRRRTHSA LKEAIGAMEH PAIWLWYPWR MNPEPPTPH MPQRRALKNV HGAVFNDLTP VQKKRQEQML YGVNIPETRQ MKFEEQHPLL AGALRKLEGQ PKGFPFWYRK Y PTRRHAYE YRFSIPVEML DGYNDDVKKA LSKGMMSIQE KQFAQEAMYM ERYAEHDFDT TSPAVLAVKR ALKCRVLRNH LL TNPHNNI IKTVLANTER KLNHALRRLR KVDFKKYWEI IRDHDVQDIL QPPNLVTYRQ GSYWKYDWNA GLAISTNLAD VMD PRGLNG CVETGRSRSE VARDLGLSYT RPLHENEKKQ LSHQAVYYER LAKFKMEQPE AARAMERERF VRKFSGMFVK MDIR SGAPD FPSTYRRLLG TKVVRWASKR HGPN UniProtKB: Uncharacterized protein |
+Macromolecule #6: bS16m
| Macromolecule | Name: bS16m / type: protein_or_peptide / ID: 6 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.077805 KDa |
| Sequence | String: MQVTETVLAR AVIKRRSPQL WGAPGAPIIR MRGHHVVWKF QSYDLVVEHT HKRRNSDIRL LHYLGKHCPH PQKSLWSPDT PVAQDRHLF MLTTVDIDAF KYWFGVKRCR LSMKPWALLA KAGLLPPSLT QNSKIMPKPL FDKESLMRYY LANRKDEDVM A REKYLNYE NSMVKTEEER AAERPVAPYL UniProtKB: Uncharacterized protein |
+Macromolecule #7: uS17m
| Macromolecule | Name: uS17m / type: protein_or_peptide / ID: 7 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence; C-terminal PTP-tag Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.012414 KDa |
| Sequence | String: MLRRSLLAFP WWNFQTEHRQ RCVLMYGGAR TKNTHNANHR VFIKKYKRNA FPNRTRHHWA VSMTGVLSQR PRRMPWPYDL TSLIFNQPR QGSDKIGYVV GTSMLKTAVV ATNHMVYYPK FNQRVSRTKR FFAHDEDLAC VEGDLVHIKQ CRKISKYKHY Y VFSILEPN ...String: MLRRSLLAFP WWNFQTEHRQ RCVLMYGGAR TKNTHNANHR VFIKKYKRNA FPNRTRHHWA VSMTGVLSQR PRRMPWPYDL TSLIFNQPR QGSDKIGYVV GTSMLKTAVV ATNHMVYYPK FNQRVSRTKR FFAHDEDLAC VEGDLVHIKQ CRKISKYKHY Y VFSILEPN VEGRERLKLG LKAVPPPLFG YPVSRRIVKL NLTSTEGTQE KLAAAIQEHV QDAYRFSGPT PDQPRNRLAD PV TFEDANN MIAPNAPAAA ALDASDSPPL LDRGEYTEVE QDTRNKKGDD YWMNLQPKEK YDFKSFKKSP RPLEDQVDPR LID GKYDIP TTASENLYFQ |
+Macromolecule #8: bS18m
| Macromolecule | Name: bS18m / type: protein_or_peptide / ID: 8 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.988691 KDa |
| Sequence | String: MNRTGGSIYA HALSQFAVCR QPWNEYIGLL TKQDSTPYHT EPQEKPAYRG RKQGREGWLF GQQVQLHYHR FPDEQLITNL SRWRTGETV GDIAMQQFRN AQPFDIEDKD PQGVQRPSPE VYMKLNYKNP ATISRFLTRT GHMYPADILP LNPEAVVKLR V AKAQAIRI ...String: MNRTGGSIYA HALSQFAVCR QPWNEYIGLL TKQDSTPYHT EPQEKPAYRG RKQGREGWLF GQQVQLHYHR FPDEQLITNL SRWRTGETV GDIAMQQFRN AQPFDIEDKD PQGVQRPSPE VYMKLNYKNP ATISRFLTRT GHMYPADILP LNPEAVVKLR V AKAQAIRI GLYPRFGNPF WFRSQKFRPK AYQENYDPTT YSTKRTVEHF AYNWVQTDRI RRYFRELEAV HTSGSASARG SG GGTTAEH KQQDQFYSPE NQPISLHRNN ISYMEDVGRS VKNPTVPGLM STKGMKKKFH NLYSSTSTKR MGFSNPTLGI KKV |
+Macromolecule #9: mS22
| Macromolecule | Name: mS22 / type: protein_or_peptide / ID: 9 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.064141 KDa |
| Sequence | String: MLRRAYIQRR YPFNKRGPRE HKSWKHHVLT EPPKPLQWRD PKVWTRDLSV MKSFDAPQWD LWQSRPRSED MDEALQPFMD MPKSLKDRR YDIPWWANPF GAWYLQNILS LELLKLKSKT NAEKIATYRS YMRSLASGKD NTMSDDDVIR NIIKERWKTL E FGDRNAGY ...String: MLRRAYIQRR YPFNKRGPRE HKSWKHHVLT EPPKPLQWRD PKVWTRDLSV MKSFDAPQWD LWQSRPRSED MDEALQPFMD MPKSLKDRR YDIPWWANPF GAWYLQNILS LELLKLKSKT NAEKIATYRS YMRSLASGKD NTMSDDDVIR NIIKERWKTL E FGDRNAGY PCTFGDYIQF LNEWFKSLDE EGMQRLREHF DRRIRPLLAV MSPVDILWLE ALTQNSPHNK EQLQRKIAFQ TS LGTPEFF DMSKRLRYEI NEDYKVRDEL GPELFALWSK APERWPPERL SKMYGLDFTL VRKILVWHHF KACYDACVEP DWS LPKRLF ALEWIRDVRA RKHGLFYGKM RFAEQKITFY SDRFLFRDLV NRREASYANV WEMDDPYRFL QTEQDYEDYW GDNY DVYRR MFPEMIGRTG EPVQQYGQMP IWAGPHRQHA NKSEHNWMFA EIGVNVGHEA LKKLELDPTN EKRRRFVIRQ PDGTL RSAK MSEMRAWYWK EEWADFRFWA PQMEWGIENT PSQEQYQEHV PDTTDADFRK QRRIQSRPVK WFYESHYTRT GSFAGF QPL RFMQRRTERE VRWPDVINAA VQIQKRKPAA YIFKAIPEL UniProtKB: Uncharacterized protein |
+Macromolecule #10: mS23
| Macromolecule | Name: mS23 / type: protein_or_peptide / ID: 10 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.358805 KDa |
| Sequence | String: MRQCVVRRYK MPKNMGVAPR FDTWNEKYEP WEHMKRMGRL VGTGFYIPPE WYNHFRMFPP INHNFQQEKT LNPHNASEPT QDDTSTLSP ERVALRDELA RKSRLVASEG MRYYNIFWVR KPLDTMEKEY YELKRRGVDH GEAIRKVLQG FYSGLAVKKR V AAIQAEEA ...String: MRQCVVRRYK MPKNMGVAPR FDTWNEKYEP WEHMKRMGRL VGTGFYIPPE WYNHFRMFPP INHNFQQEKT LNPHNASEPT QDDTSTLSP ERVALRDELA RKSRLVASEG MRYYNIFWVR KPLDTMEKEY YELKRRGVDH GEAIRKVLQG FYSGLAVKKR V AAIQAEEA KLTGRFITMR EATVVLGVLA KLHKEQLTPH QVSLLAKEQG ETTQSGAKLT AIVSRTQPHV NKEASSPATS EA VGSSTEE SLSADALASM LSEDGEQSAV GTRYQVEVKE TANDSVRQLR EKAEDQTGFP DWYTGESPTY SGTS |
+Macromolecule #11: mS26
| Macromolecule | Name: mS26 / type: protein_or_peptide / ID: 11 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.136684 KDa |
| Sequence | String: MMRSSSFCRR QIRPYYNLPS KSEHGRKMTG FLTPYRHWMW KQNELWRNVH EAQFEHLRRV YKRQWLESFR VNADEYIYKY NITKAAQLA QWECEMKEQE KKRIEARQMM DGRQALKKKH LDLLREFHER QFFFWYERAS ERLQNMNLIN YVPHAQLREH I DKELDKYV ...String: MMRSSSFCRR QIRPYYNLPS KSEHGRKMTG FLTPYRHWMW KQNELWRNVH EAQFEHLRRV YKRQWLESFR VNADEYIYKY NITKAAQLA QWECEMKEQE KKRIEARQMM DGRQALKKKH LDLLREFHER QFFFWYERAS ERLQNMNLIN YVPHAQLREH I DKELDKYV AGKNEPYPLN FVGQMPFLED GDGNIVEVPE SLLSNHMAEH PDSTAKPHEP HTSSSISEAA AFEERMLRAM VS AKEEDLK EWLGDDSRAL SETIDDISRE EEEREADIRV ARSMEETDAE REVSRRAYIE RGKTGSRSIF RPPTVSEGAG GTP SAPAGD ANTPMRRRKK GKLDRVHALQ AHQDELLAKL SSQGLKEGVD ASSVPERGKI VQSRGRIRDK AVIPTHEVLM QKPE LAAGS TPGARIQTKD MVDKMYHRGK YKKSGSGDKS DGEDL UniProtKB: Uncharacterized protein |
+Macromolecule #12: mS34
| Macromolecule | Name: mS34 / type: protein_or_peptide / ID: 12 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.72818 KDa |
| Sequence | String: MLRCARVALR ADPLNGGSSM TLGSKGSKLS PEPHRRRMPW TAAKEYVPGV VLNARDKMVL DGVQLLDIES IDRASQLDPL EVLRAVVAT REYNISTGKN IFQLASQATY NGRGQRFYRK EWQEGTYDKY VTLSAIDFDR DGNKGTAYGY ITFHGETTTR P VQVDFADV ...String: MLRCARVALR ADPLNGGSSM TLGSKGSKLS PEPHRRRMPW TAAKEYVPGV VLNARDKMVL DGVQLLDIES IDRASQLDPL EVLRAVVAT REYNISTGKN IFQLASQATY NGRGQRFYRK EWQEGTYDKY VTLSAIDFDR DGNKGTAYGY ITFHGETTTR P VQVDFADV PGWYMDFVEE RAVPFTGIVP PPPSIGTDVP VDPHSYRLKA YPYYDAPNPP EFVERLLKDR GVLPDTPTET AD VDKDPTT SDGSVHYDGK UniProtKB: Uncharacterized protein |
+Macromolecule #13: mS38
| Macromolecule | Name: mS38 / type: protein_or_peptide / ID: 13 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.2406 KDa |
| Sequence | String: MRAGCVACSR IPKLGIAALG STTTSMQATC QETQELRCAT TQMAPAVIFP LPPPLRGSYI DRKPTAASFN EHHATASFRH HMIASADVS HRPRHFYFAS GTRNDTGTRS VKEGERKQLK QQTNVEVDSV AYGDQLDELS ARWAAKFYGQ VTFGPRNYPY P SSRWLARR ...String: MRAGCVACSR IPKLGIAALG STTTSMQATC QETQELRCAT TQMAPAVIFP LPPPLRGSYI DRKPTAASFN EHHATASFRH HMIASADVS HRPRHFYFAS GTRNDTGTRS VKEGERKQLK QQTNVEVDSV AYGDQLDELS ARWAAKFYGQ VTFGPRNYPY P SSRWLARR FQMKKHRIIK RFRFRRYKLA AVANLPFAKM IRVGMLPELK SSKTKRGDVV DPTLSGQLVS AVKNTGEKRK GQ RTRPKSK YQV UniProtKB: Uncharacterized protein |
+Macromolecule #14: mS41
| Macromolecule | Name: mS41 / type: protein_or_peptide / ID: 14 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.421469 KDa |
| Sequence | String: MLKRTLTVLD QTYGPHKSYK YTYMPDPRKL APIETTQRSE IVPQSIRPPT SYVPNHETFL ERADIHRLRP TSDFKASFKD WNDLFTCDK RQLRVRGIPR MTRDAIRTAV QAFQNGNPPE RFDTKEEWLY YKQFKTVDYS YRVIPELPEK YRPHQSGIDQ A PLPDYREI NKMPQWAECE ERRQSKKTV UniProtKB: Small ribosomal subunit protein mS41 |
+Macromolecule #15: mS51 (KRIPP1)
| Macromolecule | Name: mS51 (KRIPP1) / type: protein_or_peptide / ID: 15 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.080992 KDa |
| Sequence | String: MLRCTVVGHH RSGKARAFVF RDPSLRMMRA GSGYQQLRRM GMPMQVGMGW RKVDSFHANT QYQHAWPLLS HDDLGNSDQS NNTKNIMYS MYMPKRNKGT APWFRGADTY SVKYCEQGRY EYQRYLMINR FPSEYKKHFL SFLSNIRMSS GSATIPQEAL H WLLRMIVD ...String: MLRCTVVGHH RSGKARAFVF RDPSLRMMRA GSGYQQLRRM GMPMQVGMGW RKVDSFHANT QYQHAWPLLS HDDLGNSDQS NNTKNIMYS MYMPKRNKGT APWFRGADTY SVKYCEQGRY EYQRYLMINR FPSEYKKHFL SFLSNIRMSS GSATIPQEAL H WLLRMIVD NFNPQHVHYI AAMKTLQSAG ELDMARDVWK IMERQQTWPC TATICAYLDV CVEAGEKTWA MEAWNRYCTE LK FLEPGEV DPKPISRVPF SLTREELLYL PKWKKHFDHD PNLDVMDLNR FNRTREVYLR MAQVMLAGGE RNAFQHFFTK LEE AMLNKP TPVPEPPNPH LVRRPRWAPY EHCKSVHHSP WRLQNNGRAL ALGPPVTIED EMQSRFFSND QFLVHSVKEV LRIV LQEHK RAHPTECTRC KTEAFFYKTK DADETLKFCD DLIERLFASL GVRLSNLNTS SLLSTILEVF RVVGKESGAA LLQRA NEFL ERKASLGDAE GSRENLTASN YLQVLSGFAD ESAFVYNTKK DGTCQYKTGF DPRTTMRHLA DVVQEIAGNP HVTWAA DMH LQVVETMVGC GTMKANDYFV RNVLRQFSWD SRFLEALYVE YRRQDDVDMW AELTKRALVW TARYNAPASE RLRRLIE DD YDTIRVQTRT FRELAVFQFR DVEERRHSRD VVNELPNPWY DYVAHALPFP DRDAGYPDEY GDLGQWRAPG GPGSPVRG P GYYAPPMEGE HQRGYTAEWR DLRNPMRPPE FPTPWERKYR QYARGQHPSY DMVYAGPMPE IFPMRRDFRK PTRWDFHDI EKQGKYRTSG PY |
+Macromolecule #16: mS56
| Macromolecule | Name: mS56 / type: protein_or_peptide / ID: 16 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.306836 KDa |
| Sequence | String: MRKFCHFMAN CWNSARSHAT YGAVPLTHSQ VTSVYATDGG KVDELGLLEL VEERIFSWKL NKWEMRIPPN LPNDQKELIR QEQENLKQI LSEWRKCFGA LNADILQISS LTGVPKDVVR EKNRTWLQEE VAKLRWMGEV NKAALLRDAF MRLEAFGSRD F MFMERLCC ...String: MRKFCHFMAN CWNSARSHAT YGAVPLTHSQ VTSVYATDGG KVDELGLLEL VEERIFSWKL NKWEMRIPPN LPNDQKELIR QEQENLKQI LSEWRKCFGA LNADILQISS LTGVPKDVVR EKNRTWLQEE VAKLRWMGEV NKAALLRDAF MRLEAFGSRD F MFMERLCC IYGLARQGTF DEAFTNYITE DPVTNDIFVD ERNPFKELVA HIVRNYSQID IIYDFLGFNY SEGYRSSLRR YM EYLQCKT AENVRASGRL VTGDKGEHNI LFDYCVSRES LVSGDSCQGI IDFLYINGND VTLIIIASDN PWLRNRQLPH RRQ MEGIAR RVCFVLGIPP SEVRIRNLLL PPTYLDKGSI VRLNDIVFRL SNEQSNLLIP WLTNYNKELD PKDVDYTALA KTTN EEEWL TL |
+Macromolecule #17: mS59
| Macromolecule | Name: mS59 / type: protein_or_peptide / ID: 17 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.395676 KDa |
| Sequence | String: MRCSCAFLDK SVFAAKRRVI VPIHPTPNFP AHFIKSAFTT DPLKEKQKAR FSSGGEAMRE VQDIPKNLEG ERSRRDLASR GDTEFQALV EFIEGASYDQ LISGRRFKKV YDVLSENDDM FIWLCHTAMS VLNPGDMRSR LIYNHLRILA ESVASGEMTQ R TAFRFFES ...String: MRCSCAFLDK SVFAAKRRVI VPIHPTPNFP AHFIKSAFTT DPLKEKQKAR FSSGGEAMRE VQDIPKNLEG ERSRRDLASR GDTEFQALV EFIEGASYDQ LISGRRFKKV YDVLSENDDM FIWLCHTAMS VLNPGDMRSR LIYNHLRILA ESVASGEMTQ R TAFRFFES AVRSPAYREI AKRQLEGGAA TRLAGISAAA DVMRRMGLTR RPMSSYFELY QRIVERSEAM TPWGFPPLFQ FE ERLSLEP RLKFFSRASQ QSLERRRRGH VMTPHTTLHG RRIFWIPPTW NRAGRFLGPH VTLYPGMTPD |
+Macromolecule #18: mS62 (KRIPP14)
| Macromolecule | Name: mS62 (KRIPP14) / type: protein_or_peptide / ID: 18 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.0681 KDa |
| Sequence | String: MRRFCTARVS SYVKRAPLLL PRGGRRWRSG AGTTADGGGE KEYIADSFES TAGSNAYAAM ELAAEARREI HELWLSAETA LEREKRVQQ VAALIEKYKL DPSTPREADV SRGLGDAFDR LLLLCLPLGK TDAKGTDNLE RLMHLAGRNG RELSVRTIQH L FARTDSFA ...String: MRRFCTARVS SYVKRAPLLL PRGGRRWRSG AGTTADGGGE KEYIADSFES TAGSNAYAAM ELAAEARREI HELWLSAETA LEREKRVQQ VAALIEKYKL DPSTPREADV SRGLGDAFDR LLLLCLPLGK TDAKGTDNLE RLMHLAGRNG RELSVRTIQH L FARTDSFA EALAVFYTMR RCHVAMNMEA YYSMLYSLQR LEEEGWGQHF RNEYEENGAP SEQAMDFIVK GISNALLPEN KP WLGRVMF QDRNVPDRRY DTRDFDELDT AWTQRYKSGT PAGAH UniProtKB: Uncharacterized protein |
+Macromolecule #19: mS63 (KRIPP16)
| Macromolecule | Name: mS63 (KRIPP16) / type: protein_or_peptide / ID: 19 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.922178 KDa |
| Sequence | String: MLHGTPVRRA SLRYRRPYWM MFLKGVDNWK IYTVIQQPDH QRTEMLYQAW LGGLDRPYTR PKCMANQPLW LSKKRHMLRK ERLDGPETP LEKYVLEWHK KFHSFQGTER PTPDDLHTAL DLVERPLDLS YALQLLGQCR NLNNIRFAKE TFLVFLEACL R VGRRDCAE ...String: MLHGTPVRRA SLRYRRPYWM MFLKGVDNWK IYTVIQQPDH QRTEMLYQAW LGGLDRPYTR PKCMANQPLW LSKKRHMLRK ERLDGPETP LEKYVLEWHK KFHSFQGTER PTPDDLHTAL DLVERPLDLS YALQLLGQCR NLNNIRFAKE TFLVFLEACL R VGRRDCAE YALEHAEPLG FWFIDEDHRR YLQGEQTWYK LSPLDNLYYP VEENAKLNEG RKPITRLSPA TESPGSGTAI SD GEPSTDV EGETTVDDEI AQLEAELAAL EREGGGK |
+Macromolecule #20: mS65
| Macromolecule | Name: mS65 / type: protein_or_peptide / ID: 20 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.617381 KDa |
| Sequence | String: MFSESLCILS RRFRYNTKFP ALVSYNKLPW EVVNHETPQF HMHVAPHYEQ LLTLAASSPV PHIIGSKHID VPREHRLRLL PGMLYLLDG DTLPGEFTIN RVLDPTALQY YGRLSSQIVT VEAVRMLVPD DLRLLCNCIT FKGPLHLPVA PYASLASLRG A SQGGTTGS ...String: MFSESLCILS RRFRYNTKFP ALVSYNKLPW EVVNHETPQF HMHVAPHYEQ LLTLAASSPV PHIIGSKHID VPREHRLRLL PGMLYLLDG DTLPGEFTIN RVLDPTALQY YGRLSSQIVT VEAVRMLVPD DLRLLCNCIT FKGPLHLPVA PYASLASLRG A SQGGTTGS ETGSNCFTLY HFVRPNRPPK ELQLEKYYIH APCVAPLSEF ASNSDERGNW RPRLQAPKRT RRATPLPAYR PP QSYLMGL AERLAVVPGG CFGRRSLMWG HWF |
+Macromolecule #21: mS68
| Macromolecule | Name: mS68 / type: protein_or_peptide / ID: 21 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.472807 KDa |
| Sequence | String: MRRGALSLLK AGLLGHYQQE AFEARKRFEE STTYPGPIRA ATPGDTRFYS GSLESILHDT DRHYWRAVTD DPRVQHLIPL RIRFKIFTW VTSGWEQRMQ VVQIMAPKDS TIAQVKDLVI VENQSPYLCV SSFHLAIDGK ELDPQKTLGE YGITEQSQID A IEQNDHLL ...String: MRRGALSLLK AGLLGHYQQE AFEARKRFEE STTYPGPIRA ATPGDTRFYS GSLESILHDT DRHYWRAVTD DPRVQHLIPL RIRFKIFTW VTSGWEQRMQ VVQIMAPKDS TIAQVKDLVI VENQSPYLCV SSFHLAIDGK ELDPQKTLGE YGITEQSQID A IEQNDHLL HRDDERPRDW TVDEITAEDV KRSPYKEMEM QPLQNLAPRY EARPKGYFGR TYYSGMKQSS |
+Macromolecule #22: mS73
| Macromolecule | Name: mS73 / type: protein_or_peptide / ID: 22 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.360885 KDa |
| Sequence | String: MLRRSILFRM KYADLELTTR GEFPHGMKEP AFVKKLDQNI PWYFSTYRSM YHWPITGDNW SDLNEAEKHH DLHMFYTLAW WKLGEGIFG VDEDS UniProtKB: Uncharacterized protein |
+Macromolecule #23: mt-SAF2 (KRIPP2)
| Macromolecule | Name: mt-SAF2 (KRIPP2) / type: protein_or_peptide / ID: 23 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.189547 KDa |
| Sequence | String: MLTPTRRLPK ALSPYAQALR HVALRGATAF GPGAKEMELD MLRKGTLPAD YRPPVQGRWD DTIERWAYAW QFPAEEEQDD ITKSVERNA SGMQALLEIG NKLLRSPPSP EPLSGKASKL YPPVGDMSSG NTALLPPNPT YDVLEQDVSE LMAEDAVVVT S QPRLRAEE ...String: MLTPTRRLPK ALSPYAQALR HVALRGATAF GPGAKEMELD MLRKGTLPAD YRPPVQGRWD DTIERWAYAW QFPAEEEQDD ITKSVERNA SGMQALLEIG NKLLRSPPSP EPLSGKASKL YPPVGDMSSG NTALLPPNPT YDVLEQDVSE LMAEDAVVVT S QPRLRAEE LSAKYNVPMA YIDDSSEASN ASKSLALVME DVGLEFTEDG LTVVISALSR QGYGTIGRAI FDFASAMGLG PS AEMYRAL MKYASRRGDV NESMALIEEM KGNGITPRIG NWHELMYTFY KAKDYPAVSQ IVDNMKMYAN IEPNEVTFVL QLK ALAKDN SQLNSLPEAI QLFDQMENVY GFIASRPHYD AMMFHLSQSP RPEMRLRCEE LAHKMELMGI VWNANTYLNL IRSA QVVGD VAAVEKYLSR MREEGIPASI GHLTWAVQAH VQSMIRIDYD ALKEKDESPL PTWLEHLETC FGIYELVVRR GWVMQ LPFV NALLRLTCQA TILSMERTPD EAETIGRFEE QANKIWNHTF DEWQLQKDVY SYECYIALLA HQQRIDEAEK LFQEMI LKK DLSPSRRTYH CMIFMHLSSG EEGGTARALR YLEAMERAGI QVRPSLLKKI VRVNNAAGYK RDMKRRARRI MQAREEY LA RKEEGVSFGE GGKEGASNQR ADVDAEGNSI LEPLAVSPTS TLAWWEKWKR ETVSKHELFT EEGADGTPKG ETFEEKNE A LRMMGITSSF QTKDLVPQPD RQKLLPLIRR EEGEIAGSLW AMDGGELSYP KDGGGPQGWG VRLWRERQLV KREYQKVLD GYRPVPQLST LGNSVRTAGD QLDIERSGAQ TPGELSDYRN FPDNRFDGGQ LKPESEAAPA VPFSAELVWQ GEANDKLSPY KSDEEIALE NDNTFFSSLS KETEGKLSIA VKAMQNKEEN SVDVRGKGVT RRSKFDYLEK WRDMYRHGTL EVPEGPTLNF G RTPDDHKE TMAALVRGWY QRNRKEPASE EELKRWRVDE QRSSETSASR AALKKRKQVR SRHRNK |
+Macromolecule #24: mt-SAF3
| Macromolecule | Name: mt-SAF3 / type: protein_or_peptide / ID: 24 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.621727 KDa |
| Sequence | String: MLRGWHPNSS AMQVGMRHIT IGGRHSRGGF RQPLGKHPQV KQGTVEGVPR RIPGTTKVTY TNKKGRTFSF SVPVSELTHP QVTLESAAG TWREMDTSFC ELGDIEDDMP SPVDECLRGG SSLDKRLIQE VRERFVSFCR EYVLMDTSGM KSTILSTELN A GPDYEHYD ...String: MLRGWHPNSS AMQVGMRHIT IGGRHSRGGF RQPLGKHPQV KQGTVEGVPR RIPGTTKVTY TNKKGRTFSF SVPVSELTHP QVTLESAAG TWREMDTSFC ELGDIEDDMP SPVDECLRGG SSLDKRLIQE VRERFVSFCR EYVLMDTSGM KSTILSTELN A GPDYEHYD RRLRRKRHWL AIRHRFEDVR YVIWPDVVEE TARGDSAQAD VSLTNPSLTA GEMLEALLWL DAASTFCVRK VH PSDLGDK SEFLPLDLQR EVEVVACHAR RDLDFFDPSA TSLEQFTACA ALCVNHRVPF SLFFPAQDVC GDASVSTGQC IVA NAPSPH TALGAVRIMA LISEGSGSDI GKTIMFSDAF GAVTRFGILR GLSRVMSVEA FGCKDALENV NESELCIILH FCAE VREQN AAFFRRYEAS EEDSDPQQVS FLAKYQQLSQ IALARCKRLL YHPDSPRAQV MSEDGYIPLV ELQRHAEGTN KAALI HYNL GIRSAQGMRR VALGAQSSAR LAELVSRLEE ASARVSGNTL VNDLVHHLSH KAAAGKMSLT LREVNTLLPL LSRMRR ESP NGALDARFDR VFNAIDTAIG AAMRHNCTLD ELLDLAEGLA ACEMVPSALK QVEMVLIRSV MMHECSPMHL RRMLQAM FT LMRTSVPQVL LQSVASRVAD YIKEASHMDS SSSNGGGDEK VKNHEECEQL LELLVVLGKC GYGALPGLVT IYWEAQLI D SMQLNPRLRC SYASLLASAA FALKKHDKRA WEGLADESHR LFMEYTRCNK ENDIGRFAEC VTGLAVLTQI KDNTNSSDV AFLKEYLSAT SLELKSCEVI RVQELTDLLG RTLEWSEALG VVAPDVVIQL EKALFVMLEN VSHTAPGVGI PDELVTAACC LVDMSSASL ELRKAAAGVV GGAIVHAEEA LETLRSGAPT QVRPGHSFDV AALASAEREN VYKNSILQYC AALQRSGMST H VEELWS UniProtKB: Target SNARE |
+Macromolecule #25: mt-SAF5
| Macromolecule | Name: mt-SAF5 / type: protein_or_peptide / ID: 25 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.949414 KDa |
| Sequence | String: MRHTIPFFRR SAFVPAPGSS LLNPRSQRAK VRRMVAAQKA QGENFERQAL YAELGGSPSA RAPRSKGERS KEATRRVGCE VAERAKHMT DAEWEGVPVD EKHAFAKYMH KVLQEHPTET TEQQRRRYFE TTMADVFELD PRKTVRDEYE RVKLGLPVHL K NPQYSLGV ...String: MRHTIPFFRR SAFVPAPGSS LLNPRSQRAK VRRMVAAQKA QGENFERQAL YAELGGSPSA RAPRSKGERS KEATRRVGCE VAERAKHMT DAEWEGVPVD EKHAFAKYMH KVLQEHPTET TEQQRRRYFE TTMADVFELD PRKTVRDEYE RVKLGLPVHL K NPQYSLGV SQAVYDAADA SLFDPENVHR LENAMTHVKQ VFADYVHKKR EGVSTEAERR MLANLTAELN LETQKHLANM FK YAEMRLR QVKLEERHHQ LAEIERLRRM AQQRGGVKGR KGGSRKMSRM ERLKRVINRA VGLDIAVAET VLTEMQAQEE FLQ FCEVFA RLTLGSGFKH TGKDENLSAY IESLRKLYSM DAATLSTLDV VQYYSSKEGA HPVDWAKRWY ERALLLPLQS TPEY QKLLQ IQQRDESTVK HIKETAGTGH AFACEAEAEV ARIKTQKVVN LVEKMFMDPK DKRLESLHEK RLRYLAHMQM ERQIR CVRE NAKLFDGVEN MPEAAQCREL YEKIMEKKTA QCNMTSPPEG EGSAIQSAKT EGDHCEVGPG MFNVYDDAEA SSLFEK IRE ITLRVIRDRR VQSAAATKAR MLNRIIRSLK GGERSIAEEL RALHQQRKEK MTMRILGIIE NDVKTEMEWL QNMEEAE RP PLLPIPENMS YVSAADVQAW RELREDDERK AANPFERRRR TFQPELLGQA WSVPNKPLLF WGTGVSAVQQ ALRHVAED A ERKRQGLLLA PPYPCAENPW GWRLAKDILD DNN |
+Macromolecule #26: mt-SAF6
| Macromolecule | Name: mt-SAF6 / type: protein_or_peptide / ID: 26 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 76.781398 KDa |
| Sequence | String: MYKKALHSFF LKVHPDFFHH NRSQQTVNES SVARLNELLS WAKAFKSGHL QPPPSSSFTL TFYRKPDDNM GNGETRREGS GSPVSSLVG RSMGDGSLSP TIIQSTFELP SNFAPSDNHR GTVERAVNKF LRDLLRRAAC IDSVTESISE AEDATAARAE A KPLRRRPR ...String: MYKKALHSFF LKVHPDFFHH NRSQQTVNES SVARLNELLS WAKAFKSGHL QPPPSSSFTL TFYRKPDDNM GNGETRREGS GSPVSSLVG RSMGDGSLSP TIIQSTFELP SNFAPSDNHR GTVERAVNKF LRDLLRRAAC IDSVTESISE AEDATAARAE A KPLRRRPR GSQHRGAPTG PKSLLDEAVE SMTVQWSLTP APTLQELIEA DQILFSRDLS PLQSAAALST LQRHLGELNY SA WESMPVI VSNQFSIGDL TGTITIPWDF TPEQFHSFMA HNEKGVARCR EVAIQYASTI EQLIAELCTA LELDDILVSC SHQ DALRLM ELLHRNRELL IQYGLSKLTL EVGNRHATRA NGVVIINCSL TSEQLRPWLK AISPKLPLQQ RLYELSKQML ESTL WHLKE FRTMVEPGGV DAFSNDCTYA ERLQWSKELF RIGPSLAPWD WSEMTFVLSP DVDIDWANGL LALPYNFDGD ALVRY VEEV QQEAKSRKRE ELLAASAMQR EEEERRRKQQ HDEELMVECQ EKGERSGDDS PTVAERMRHL YRQTNPHMDE YLASSN SRV DTLPVERPLS HAVTFNSDAE AEDQLKWEGF YAEPYVDQAP TSDIDDMAHT FMLTNRWHRE EAAKKMLDQL RGTYGKK SR RFEYQKMGDV LEINNAKVQP KGFPTLTRGI KPGC UniProtKB: DUF4460 domain-containing protein |
+Macromolecule #27: mt-SAF7 (KRIPP10)
| Macromolecule | Name: mt-SAF7 (KRIPP10) / type: protein_or_peptide / ID: 27 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 76.675156 KDa |
| Sequence | String: MLSHAVPRLR LAAAAVKAAE WYAAENRRKQ KGGDVIREKV DCTAPLWFDD TEQNLLAREQ HVRSPAYREL HRPALTNAAM GLYGGEPGF EKAHRVWVDP DRPHIKHIYN QTALARNLRY ARYGYFKRDM HLLDVDKLVR HARLLPTPGR LLTDFLYQRV P LPDKSCAA ...String: MLSHAVPRLR LAAAAVKAAE WYAAENRRKQ KGGDVIREKV DCTAPLWFDD TEQNLLAREQ HVRSPAYREL HRPALTNAAM GLYGGEPGF EKAHRVWVDP DRPHIKHIYN QTALARNLRY ARYGYFKRDM HLLDVDKLVR HARLLPTPGR LLTDFLYQRV P LPDKSCAA LIRYQRQQIE MLEVWGRHAS FQCAVEMFER MIVTNIPPVE VGVETHGEMV LCAAACGKWE EGWNVYANRA RE LEKESPE SFILNTFFFD ALLTLCVAAG RVSEGIDTLE EVIKRNLRPR GTMLNKAMIL YSILGEQMSK HEASRATSEG ERS YLCEPE EVEKMGLEVW SLFDFYQLPR TTASIEAYMR MCCAFNKPTL VLKAQGFADA SDIRLSIECF HWLVYAIRGV AGFG DYVMD VLSQLRPRGL TPDFVLFTLS FMYCALQRDG ELALAIFDQH FVHQNMNPTP EMVLLFIQAC SNCEEPTAVM LERSE TLIK RLEAVGSSVD LISPIYDQFL ELCAHLGAVA SGFSALKRIV GFGKPLTTRM INSLLLANSN AISSNGSLSM TEELVG FFT LLKIRPNADT EICVNLCRDA FGESPVVNDF IKVIGESLQG DSEKGEAPQY DEDIPVIQVP PHELRQLRTE WKLSPRD IV LRRFGQHTKP PGKAALDVGS MRGSVIPFGR SPGEQLV UniProtKB: Pentacotripeptide-repeat region of PRORP domain-containing protein |
+Macromolecule #28: mt-SAF8
| Macromolecule | Name: mt-SAF8 / type: protein_or_peptide / ID: 28 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 76.72518 KDa |
| Sequence | String: MMHRTAARLR MPRKPTPYVR KFLEGCPLPE TLVDDIAGAN LKSMAPFFTT APRYIVAAES RLSKLFFHHA LYPAGGARRP CRVLIVRGG RSVREPSFTI NTGGGRGEVG GGSRGYRDPA RRAYFYARPS TVGPFYSGNG GVSSNVAKCH SGVGSEGAGL V KRASVDGL ...String: MMHRTAARLR MPRKPTPYVR KFLEGCPLPE TLVDDIAGAN LKSMAPFFTT APRYIVAAES RLSKLFFHHA LYPAGGARRP CRVLIVRGG RSVREPSFTI NTGGGRGEVG GGSRGYRDPA RRAYFYARPS TVGPFYSGNG GVSSNVAKCH SGVGSEGAGL V KRASVDGL LSPLCGVIEA HFAVGGTCND AVATEGDGTE SLTKGGEKLC VTLAGLLSGG HGGLMMDDGN CASNVRAAKR VA RLLHDAA HHLSSFFYVH TQLPDSALFV SRPEASIASD GKGDGLLPSH LAREKDNDGR PSVEGVAVFR LAGGLEPTVH FAV GAPLSV LQRGVDGTAS RGEKNSAEEG LNSAASTTVL PFGHIQCLLR VRTRGGKHCA AGKEGSEGTS NTPWCNTAGN DDIT SNFAA SGPQIAGGIV EPWKLGVSLD PKVPFFMRTL TEKRPSFSCG EGYLGTCSRS GDVDGNTVND VSSSSGGSRT RAPGE VHMN HLLVRNDCET YLLPQRELLL SFHVPEEAEA MCKEQNEERM RRQAALGYGS PSHVFAEGPR TFARVLHGMK ANLAAV EEA SSTFRQGAAE GISPQVNGGS TTSGSSRVYE VRALPGDVVF VPRGWKYSVE RIVGTAIIDA VAASTASPRE ALRAVFR TA PDPPLPQDVV RCDEGHAGAG EMPGGDSSNA EIVGVEVDAF VLCYKPYPVL SNAQASTYVA ANYVHSGIDD FYAKGGND V YHKYT |
+Macromolecule #29: mt-SAF9
| Macromolecule | Name: mt-SAF9 / type: protein_or_peptide / ID: 29 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.781789 KDa |
| Sequence | String: MTGPTRALFL SSGINLGRLR LAEQFSSMNG WQSKEDPAFD AYVKERRRKE NYEAFDQRVE RGYAAAAKLH KAEIQNAVKR RLKSSGAKF TAETLREMSS AVTERLAWLR DVWAQIDADY RSGDSARQET AAQEISAALR GEPNDYMRWV YETKRELRFA G PVGRRAIQ ...String: MTGPTRALFL SSGINLGRLR LAEQFSSMNG WQSKEDPAFD AYVKERRRKE NYEAFDQRVE RGYAAAAKLH KAEIQNAVKR RLKSSGAKF TAETLREMSS AVTERLAWLR DVWAQIDADY RSGDSARQET AAQEISAALR GEPNDYMRWV YETKRELRFA G PVGRRAIQ EELQAAELPE VLDEEVNRYH DLKLNMMEIE REVKAKYGVA GQQHWAELQA AKDEEYIQKL DEAAEVYKQL LD QSARLDE SRRSELQRSY VERVHQAQVR FKAAMELEGQ REQLIEAHQA MKEERMRTER EKRRQLLREA AELRAQGKKS ADV LTALKE RQLDANAKRQ AEYELKECED ILKRKSEMLD MIAHFKHDVE EREGREMLQR QKSDEERQVN VFGFYEEVGV EDGL SISSE GTTSQGGSSG LGTVSTSTSC AKSADSNSSA QPSQKLRKEE LWKVINADTY EDPFRTVHQA RLDAVKTYDP AYART FPLN LVLGRKYSRQ GAGEMAAGNE TDKQILQKGN NILYSFQWGL NNGTVHDLDA DGGTDYFMDG AFHVRDKETG DIDWRY EKK RGGPVFRGPK FYRLGAQREA ADPGERAMDP TPYTSTPREH KWRSS |
+Macromolecule #30: mt-SAF10
| Macromolecule | Name: mt-SAF10 / type: protein_or_peptide / ID: 30 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.306758 KDa |
| Sequence | String: MPSASSGYTF ADFLRRLERS PDSHMAPLYH EHRELFVRRH DMFARVISSV TWSKGVALVA AAGYTQAVNV TIYRALLARM LLHNRHVRQ CGAGSVVPWS AALRTYSEAI ATHGNAVPTR MTLSALRLCT PARQWVAAIS LLMLSQANDK LTLPMLIDAA G CCATPAAW ...String: MPSASSGYTF ADFLRRLERS PDSHMAPLYH EHRELFVRRH DMFARVISSV TWSKGVALVA AAGYTQAVNV TIYRALLARM LLHNRHVRQ CGAGSVVPWS AALRTYSEAI ATHGNAVPTR MTLSALRLCT PARQWVAAIS LLMLSQANDK LTLPMLIDAA G CCATPAAW EKAMALLGRF HAQSLQVLPD SIQSLRPVGT SASTVDAAAH ALLPRSEGPT PEQKHILTVI NKVVSAVPWQ VA LSNEMCR SYLTHLVAST TLRPTEKTAS LTTAVQQLPW EAFVTLMKTV TATVQEGSQG VLGSRSTPQL PPPQDGMERG DVA KSLLSN SIIREGVNLL QSEPETAIPF ITTILYKLPS AEAAALFLSE ATSAYRNSSS AVVAAAIRHP VVVGALLKRC ADSN SWYLA ASIFKSTSPT AIPCDVASDL VIQMRRANQA PLVVDVLQKY IVPSRTKLTE EAIEAALLCV LVHNRALAKA SAVVA GTSP DNRTGKPNGI GVANGVHWIS ALSWATDLLE EGVESRILQT GTTPSVGGVN HEDPTVLLRK KTLSPRILSL LIYICV NAG SPRGGLFALG YARTVSKTEL ELSEEITALL YCMMYDRPRE AESIIQHAVK KHGEYKGKYL GRLLVASQEA KGSALRN QT UniProtKB: Uncharacterized protein |
+Macromolecule #31: mt-SAF11
| Macromolecule | Name: mt-SAF11 / type: protein_or_peptide / ID: 31 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.916957 KDa |
| Sequence | String: MRRLSVSFLP FHLIVTTQRR QFGGNSKDGH KGEDKPIERV GEVPPTGPHP FNITSGAEGQ ISNQQRKAHD AITQLFRDDA QRKALYQKP GRTIGAQTTT AAISTPPNED EFEGLQPISG DAEPSSITAV EAVEQELLDD AFLYFPPAQQ SASGSVATTA T NELYVPGQ ...String: MRRLSVSFLP FHLIVTTQRR QFGGNSKDGH KGEDKPIERV GEVPPTGPHP FNITSGAEGQ ISNQQRKAHD AITQLFRDDA QRKALYQKP GRTIGAQTTT AAISTPPNED EFEGLQPISG DAEPSSITAV EAVEQELLDD AFLYFPPAQQ SASGSVATTA T NELYVPGQ QIIPPGLTRY RVDVQYQGND FDGWWKSTTR QLFRREVGLD GSITRVPVAE PGTIGNAAGA SRGETMGSRY HA RTVLEEA LAVALDVNTV RVVAGVIPEV GVSVRRLCCH VDVPSHIELQ PRTVIQRATM WMEKRQQPLA ILSYRRCKNQ DFH ARHSGL RRVYVYRILN RVAPPLFDAG LQWHVDRHLD VDRMKRFAKA LEGTKDFGYF ADPKMANALR RAAMSPGGFS TGAV TEENF QPKATGESHR VTRGKAPKVT MEKGPSNLDR AAALPTFNEY GQRVVQPGAH GKEYYRVATN LPTVRTVDRL DVVRQ DDEV LIWFVGRSFL RHQIRNMVSV LKAAGHGLWN DLELQQALQS GFEPSRHRFK RERFPTAPAY GLTLWDVEYP DQHRDD YVQ FVDSGPYEQV NIARDI |
+Macromolecule #32: mt-SAF13
| Macromolecule | Name: mt-SAF13 / type: protein_or_peptide / ID: 32 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.690703 KDa |
| Sequence | String: MKRYRSSVVT RLVRAITLRP DATVDPERYP LGYVPLDGSE SVDSVWSLVK SGAFVAPLSK IETIHRAHVG IRYLTQSEYP ALSSIDVVG LQTRLKELCS RLLIRRDFWV LDDYNDPELN SSFGIQNMYF DNFKWSQVLW RRFQQYVEEY FPVAEHTHLT Y DEYLQLLR ...String: MKRYRSSVVT RLVRAITLRP DATVDPERYP LGYVPLDGSE SVDSVWSLVK SGAFVAPLSK IETIHRAHVG IRYLTQSEYP ALSSIDVVG LQTRLKELCS RLLIRRDFWV LDDYNDPELN SSFGIQNMYF DNFKWSQVLW RRFQQYVEEY FPVAEHTHLT Y DEYLQLLR SFSHFEQGAK LLPLLPKRYR IHPPFGVPAL SRIDMEPLLL YSQWLKNFRG PLKLDAALVI RSGCGAAVFA TK LNGVPIV RGVDPNPRAV MSCRKDAQRM GRRFDSISFR VGEMFPDKDD GNGVPNSRKY DIIVFYPDQG CYNLFFTNAI GEY APVLTG FAGTLEHFFE EAGDYLSDSG VIVLCCTNVY SILKPTEPHP IEYEIKVNRR WVLLDYYDMP VRGKGTLSHT PTDH HYRIP MEMRKCMRSE LWVLHKMTSI AHFAHIHNIP GAQPPSCVVS HWRNKAIGKL RRAVLKGQVE SSGGDWDDYK KRLVH LLQE QSENDEDDKA QAIRMAMDPN YPKELADRAR AAIEKNMDTD KAFHNNVAKA FGDISPRERF DAYLSCYRL |
+Macromolecule #33: mt-SAF18
| Macromolecule | Name: mt-SAF18 / type: protein_or_peptide / ID: 33 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.508039 KDa |
| Sequence | String: MLCHNRLLLV NKQTQKYRTK LRYRFRQPSV VPLRQTLQQR HNTILEVLRR RRINSGDQSP YRYVEERLYS KPSRLDREGV KVNKTYALQ GLGDLEPLRY GANFGISEKD ALKYETVAEK AKYMEPPIPY SSLAARKLAA GALWPAAPDP EGMISKEVRL L RHESSMSP ...String: MLCHNRLLLV NKQTQKYRTK LRYRFRQPSV VPLRQTLQQR HNTILEVLRR RRINSGDQSP YRYVEERLYS KPSRLDREGV KVNKTYALQ GLGDLEPLRY GANFGISEKD ALKYETVAEK AKYMEPPIPY SSLAARKLAA GALWPAAPDP EGMISKEVRL L RHESSMSP SARAFSERVA YHLRRSLKAC PGHIAEHIDF TQLIIQEVLG SRRSKEIYIV WFTVDPGARF ELEPRLHQLN HW VQQLIIK RVKRRPHIPR VTWIYDGGRL ERELPRDVKQ ELQSFVADAA TTLESRVKYL KELDTMNQRM KDIPWFMPYL WSK EEKAAR QKSMLADLEE VERRKNEHSS GRSAPPRTSP PPQFVR |
+Macromolecule #34: mt-SAF21
| Macromolecule | Name: mt-SAF21 / type: protein_or_peptide / ID: 34 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.610793 KDa |
| Sequence | String: MSAIKRSLNV GVVGMGNMGV PIARNLGFKA RSAMYLQIHS RALSKAKRVC DDLSVDGATC AMRIHDRYST MTKWCDVIVL ALADVKASR HALLEDNESL IMNARKGQII VDHTTVDAET SRECAHEAAR RGAYFLDAPM SGSPRACFNG QLLLMVGGDA E TFQKMLPI ...String: MSAIKRSLNV GVVGMGNMGV PIARNLGFKA RSAMYLQIHS RALSKAKRVC DDLSVDGATC AMRIHDRYST MTKWCDVIVL ALADVKASR HALLEDNESL IMNARKGQII VDHTTVDAET SRECAHEAAR RGAYFLDAPM SGSPRACFNG QLLLMVGGDA E TFQKMLPI FHMYADNVFH MGESGSGTAA KMISQALVAS HNAAAAEALS MAHALGIEDQ SKLVQVLDAS WGSSTMLRRN AP LLQDLIR NPDKTPPTSA VSVDRLLSDL AHLDASLPKR GGEDEPFPVF DAALRSLAAA SDAGMGDRDL ASVVHYIEAG EAE LRNRTH VPLGGEHLTG RTAATAANSS VNETPSTVNG SSEVPNEFGV EDFY |
+Macromolecule #35: mt-SAF22 (KRIPP17)
| Macromolecule | Name: mt-SAF22 (KRIPP17) / type: protein_or_peptide / ID: 35 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.132621 KDa |
| Sequence | String: MTDPVRRDPT EFLRVLRRMS RTTSWQKTML FASKGRMVGY RLTREHYNTV LFSQSLWGRA LEIVRVVRAM QEDKVQPNGA TYYYIVNGM GNADHGWNYD FRINRRLEKI QHWRVALEAL EACEANGFDS TDTMHNSALI TLVIPGFNRW QQASLLLQRM L REDRRMHP ...String: MTDPVRRDPT EFLRVLRRMS RTTSWQKTML FASKGRMVGY RLTREHYNTV LFSQSLWGRA LEIVRVVRAM QEDKVQPNGA TYYYIVNGM GNADHGWNYD FRINRRLEKI QHWRVALEAL EACEANGFDS TDTMHNSALI TLVIPGFNRW QQASLLLQRM L REDRRMHP TMVKFYHDCL VRNNRPREAS SLMRLAAERG VHGYEDKWEA DVYKGRPLDS EVMNESEGNG LRRQASSLAF AS LMLRGDQ RPLPENLQAL LEEETTRNIE AERSVPVPFS AGLHATEINS VFRPRVYRQL WYKWQHIANR YRPTAALKRR QLA PRDSPT GIPGFYRI UniProtKB: Uncharacterized protein |
+Macromolecule #36: mt-SAF23
| Macromolecule | Name: mt-SAF23 / type: protein_or_peptide / ID: 36 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.240164 KDa |
| Sequence | String: MVAKLQHNSA PTTLNFYEKS FQQLSDVQQR QTGLLIGAAV GDAAARALDG YTAEEVAAVA AESGSLQDED EDPVVFASVT PREHKSGLL RHHSYTFYLF SQLLRVMATS RGDFPVQYVK NEWVATARAH PDCFVREHAS LLHVLCITMQ LPVIYPWADD S TLREYASG ...String: MVAKLQHNSA PTTLNFYEKS FQQLSDVQQR QTGLLIGAAV GDAAARALDG YTAEEVAAVA AESGSLQDED EDPVVFASVT PREHKSGLL RHHSYTFYLF SQLLRVMATS RGDFPVQYVK NEWVATARAH PDCFVREHAS LLHVLCITMQ LPVIYPWADD S TLREYASG FLEFLTETPA EQAVASREDV YAYTNSVLGV ALRCLQSNPD PYRNAAFMAA PGTAHVFPDD LALYCPPALV GS SHSRTEE LSETDSETVP LFPARLLESD VRVVRECLVV ARGAASFAEG IKAAIHLGGP VCQRSLIVGA LLGARMGVRR IPI SWLSAT YDHVPLVTLA LQVAQWSWNP PHH |
+Macromolecule #37: mt-SAF24
| Macromolecule | Name: mt-SAF24 / type: protein_or_peptide / ID: 37 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.648047 KDa |
| Sequence | String: MRFINQAASD RAVINAGGRR FETLFSTLHR YPDTPFAQLF PLPGRGARQH RGREFFLDVT PHVFEYILGF LRTNQLNLPA ENLQIRAEV VYSMNQWGLL EHAFPPEVIE DGEGCSTGGA VVKLPDVCVV QVCDHMQHDQ GVKRHALTIT YGADGFQLRS L IRRVRRDL ...String: MRFINQAASD RAVINAGGRR FETLFSTLHR YPDTPFAQLF PLPGRGARQH RGREFFLDVT PHVFEYILGF LRTNQLNLPA ENLQIRAEV VYSMNQWGLL EHAFPPEVIE DGEGCSTGGA VVKLPDVCVV QVCDHMQHDQ GVKRHALTIT YGADGFQLRS L IRRVRRDL ERQLSSTYWQ CYQTNERAAF FVTTKVANGT ADLLTTSVTQ QLVEHTESMG YSLASSYVTL SPDVVHTSVR ML IHNFTFR RSRRVEVEPG DGIALGEGSE TIEAEPNIPT MHVGPRREPL NAAESIPPRN ERAVNIWTVD UniProtKB: Potassium voltage-gated channel, putative |
+Macromolecule #38: mt-SAF26
| Macromolecule | Name: mt-SAF26 / type: protein_or_peptide / ID: 38 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.538557 KDa |
| Sequence | String: MSTSSRGILI AGLTNRARTQ MAEGILRRLT AGTIFIKSGG VHHESTIHPL AVKVMKDIGI DISKQSVTSL EAARRQQNTY DVYISIDCS YERRTLDKSQ PPRAEVRRRE SFTVEYGDDG SGPRGGDDPV RYDPLLVPRT PSHWSVGVDA TDVRRRWQIW S PRDPSIFH ...String: MSTSSRGILI AGLTNRARTQ MAEGILRRLT AGTIFIKSGG VHHESTIHPL AVKVMKDIGI DISKQSVTSL EAARRQQNTY DVYISIDCS YERRTLDKSQ PPRAEVRRRE SFTVEYGDDG SGPRGGDDPV RYDPLLVPRT PSHWSVGVDA TDVRRRWQIW S PRDPSIFH ETSTRKFQDH LYEGEPLFMQ LRQCPWRTQA KVSERWEMDE VAIRYPVERH SAHELRFVQA REQLLQRSFD LL TKLEKHY GERLLLDRAL LEEFIS UniProtKB: Phosphotyrosine protein phosphatase I domain-containing protein |
+Macromolecule #39: mt-SAF27
| Macromolecule | Name: mt-SAF27 / type: protein_or_peptide / ID: 39 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.936748 KDa |
| Sequence | String: MHPRLSGQGV FPRYGGGWMH QFGHVYALQQ ELLNAEANNW FRAMELWHTA RHEGVAMNVS HYTNILRQCV PPTAWEASLM VLKQMRREN IRPDVVGVGC ALAACADANR SEEVEKVFSC FSGKLKLDSI CYLALIKAKM SRGGYAEALA VGREQDADGV P FLPHTYTH ...String: MHPRLSGQGV FPRYGGGWMH QFGHVYALQQ ELLNAEANNW FRAMELWHTA RHEGVAMNVS HYTNILRQCV PPTAWEASLM VLKQMRREN IRPDVVGVGC ALAACADANR SEEVEKVFSC FSGKLKLDSI CYLALIKAKM SRGGYAEALA VGREQDADGV P FLPHTYTH LLEAAEVADD GEMALELARR MSSEQWPVSD RGREALKKLS TRHHWEEVAE YKQFVAVEDH KLSLPPTLPR G UniProtKB: Pentacotripeptide-repeat region of PRORP domain-containing protein |
+Macromolecule #40: mt-SAF28
| Macromolecule | Name: mt-SAF28 / type: protein_or_peptide / ID: 40 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.468707 KDa |
| Sequence | String: MWRLSRSLRS NSLHNPGPFL DGALQLIKLH LAHKNAAADK NTKACSDIEG EFLRELEAFR ACFTMSSSLK VAKLYTKKLH GALSYFQLY DDPLMRQLDM IIGKQTMQPS AGRQHGVFKA PVAARLDPFF LDEREETVLP SELPNPPKPD PSTPLRERAL K VPAQHRGH WVLRDPDIAI TREERRTDPW |
+Macromolecule #41: mt-SAF29
| Macromolecule | Name: mt-SAF29 / type: protein_or_peptide / ID: 41 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.048514 KDa |
| Sequence | String: MRRKTTLNIG QVICFSSWND GSEGYEWKSR ALSEKRSLAL EFLGNVNKRV SIHDAIRLKA DINKKAISNV SCPSFFSGIE GADEDEDQS DMSLCSLLGV LEGEIETDCI THLSPSDASL LKEEFLCDYD PSDTKRMAKW VNLRSETSDY QSYGAIPEGE R SLWSAWYL RNIKAGKKPI |
+Macromolecule #42: mt-SAF30
| Macromolecule | Name: mt-SAF30 / type: protein_or_peptide / ID: 42 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.659756 KDa |
| Sequence | String: MPPNSATRWL PFVSSDLKDY LNRYWAVMFT VGARPIETGH IRHYVSWYCT RMKVVLLDHH VYVEPLRQQL QEASRTPELP LLFVNKKLV GTLRDVELLE REKKLKDVLH FGFEWRVGGS VAATNGQKSL MGALPAPYGD AEFFRGRYRG PPVARPVVSL P TLHPFALR SEE UniProtKB: Glutaredoxin domain-containing protein |
+Macromolecule #43: mt-SAF31
| Macromolecule | Name: mt-SAF31 / type: protein_or_peptide / ID: 43 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.673535 KDa |
| Sequence | String: MNCSSTLACH AVVSAPSTAS LITSCWTPQQ HCYRQLMKSL RAAYFHDRSK LFWSRHRVLV EFYKYSEEAN EEAVKQLVAI GLEVAAFID HHMRTDVERI VKHNETMMAL PVAQAKKFRS DYLLAEKQHD SWCKQKIKNI MKRRPPPPYP FF UniProtKB: Succinate dehydrogenase assembly factor 3, mitochondrial |
+Macromolecule #44: mt-SAF32
| Macromolecule | Name: mt-SAF32 / type: protein_or_peptide / ID: 44 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.550807 KDa |
| Sequence | String: MQRSQLARKF ATQVGAYSLL RGRCHAVSTW KVMATQAVTI PMYLPVRFHS EGHAAGEEAG RTGQYLLNKD DVLTRVLEVV KNFEKVDAS KVTPQSHFVN DLGLNSLDVV EVVFAIEQEF ILDIPDHDAE KIQSITDAVE YISQNPMAK UniProtKB: Acyl carrier protein |
+Macromolecule #45: mt-SAF33
| Macromolecule | Name: mt-SAF33 / type: protein_or_peptide / ID: 45 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.291121 KDa |
| Sequence | String: MRHFGPSIAH FPLSANQLRF KASHIRIANR KRVEMFVAKR FHLPTRLATA APDIAAEWHD ELNPMHMYPA IIGIGHVQPV WWKCAICSH SYQMSVEKRV VRGGGCPQCV VNGKRAVADG ALEGEMDSQL RPKRPVMFNM RTKY UniProtKB: Treble clef zinc finger domain-containing protein |
+Macromolecule #46: UNK-A
| Macromolecule | Name: UNK-A / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.805216 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK) |
+Macromolecule #47: UNK-B
| Macromolecule | Name: UNK-B / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.315846 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #48: UNK-C
| Macromolecule | Name: UNK-C / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 869.063 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #49: UNK-D, UNK-M, UNK-Q
| Macromolecule | Name: UNK-D, UNK-M, UNK-Q / type: protein_or_peptide / ID: 49 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 783.958 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #50: UNK-E, UNK-P
| Macromolecule | Name: UNK-E, UNK-P / type: protein_or_peptide / ID: 50 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.847734 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #51: UNK-F
| Macromolecule | Name: UNK-F / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 954.168 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #52: UNK-G
| Macromolecule | Name: UNK-G / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.464797 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #53: UNK-H
| Macromolecule | Name: UNK-H / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 443.539 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #54: UNK-I, UNK-M
| Macromolecule | Name: UNK-I, UNK-M / type: protein_or_peptide / ID: 54 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 698.854 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #55: UNK-J
| Macromolecule | Name: UNK-J / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.379692 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #56: UNK-L
| Macromolecule | Name: UNK-L / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.890321 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
+Macromolecule #57: UNK-O
| Macromolecule | Name: UNK-O / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.571161 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
+Macromolecule #58: UNK-U
| Macromolecule | Name: UNK-U / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.060531 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #59: UNK-I
| Macromolecule | Name: UNK-I / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.847105 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
+Macromolecule #61: mt-SAF1 (RSM22)
| Macromolecule | Name: mt-SAF1 (RSM22) / type: protein_or_peptide / ID: 61 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 119.799398 KDa |
| Sequence | String: MFFRFHSTTL QTCLTSGVAR SSVAVAVPLR AGCTRREMSS GGDGVLEGAM HKPGESGLQA GSSTTIAGKE TWSQFSTKMR YGRRRIRVI DVAAKMSYEY QMLRKMCKRR PAMRQWAVRD DFCDMNPGVV IMSPSMQAAF MKVFRMKEKG LIRQCLRDIV P VIEYRNRE ...String: MFFRFHSTTL QTCLTSGVAR SSVAVAVPLR AGCTRREMSS GGDGVLEGAM HKPGESGLQA GSSTTIAGKE TWSQFSTKMR YGRRRIRVI DVAAKMSYEY QMLRKMCKRR PAMRQWAVRD DFCDMNPGVV IMSPSMQAAF MKVFRMKEKG LIRQCLRDIV P VIEYRNRE EPARLPTNPA DMIPEGEPDT LNQKKRELFE RREKGENFAD LRLHDKRSQA KLRFRIRQRL LKFQRQLAVA NA IASRSVL YSTNDAIGYF LFRGAAMYAG MHRVFFELSK QLPHFVPKTM LDFGAGTGTA ILVAKEVYDP GSLAYPLYRS LRQ TMQGND SSRTHQLSEL RYDLKRLQRN NEEKKKVRFM AVAALLERGE VDPADLPEDL KREIAEVATA AATAKKDRLV REAH ARYRD VVDGTEWESG DPLGEVRAST EDPEDVIDGE QGDGGDDGEA AKGRPKTWWE KLIDVENETA RTRAARRLRP LQEVT AVEP SPGMMEIGTM VLHDDVPNVT WKRYLLPEDE AIQHDLVVAA YSLSEIATSE NRRRIVQQLW KMTKGVLVFV EFANLN NFN ILMEARDWIL EEKDVGLWDW QPTIVAPCPH EHRCPLRHCK TGVKRKRMRI CSTEAHYRST FVEVWARHMP LKVGIEP IS YLILARNELV PERAERRREQ LKKAEEMKRR ERDVKQQQLH EASLAVKDVV FERLSDEALH RVQSSVPQPL TDIDEVRE A STSATSTSLL KDLKDGATST GEIGHMPTDV PRLVKTGNTR HNRLIFPLQF PPATHKFNRA FVDAGYQRQR AITPAEMLV VRQEVEQLQQ RVMRAAPKYL RVVRDPRCHG KVQADFCTPE GDLVSGRVYR RFYGDRNRVS AHSTMRWQHI GGWKLLKRIR RGSLFPHNV PLYAVTKHAQ IDFPNTLLDT KHSTVEQTAM QYNDPMSLVE MPDDGLTREE LKQKRRLQRD VELQKKVEEK L EDIFGVNA KDWKTDDLGG GRLDARREIS EQQWADAVRR AKIRTVQHTK NALPFAAKKR AAQRALQVRR RNVRLEMSGN RR R UniProtKB: Methyltransferase |
+Macromolecule #62: mt-SAF14
| Macromolecule | Name: mt-SAF14 / type: protein_or_peptide / ID: 62 Details: Protein of Trypanosoma brucei brucei Lister strain 427; sequence accessible on TriTrypDB; where necessary, the reference strain TREU927 was used to complete the sequence Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.32082 KDa |
| Sequence | String: MKRIVVPHRW SEMNRVEHPP LMMKQLFQGV CGGLRWLETK SLAQYLAVRA IEEGYPSTPG VRKALHVKRA PLQKRRVVQR STKSATAAC SLPTTAAPAQ LPLTSSGDAL TVTGGLSLQV TKQKRLVSYD VLDCTLGSGY HAGAVLENGG PYTRVVALDC D HDAMHAAR ...String: MKRIVVPHRW SEMNRVEHPP LMMKQLFQGV CGGLRWLETK SLAQYLAVRA IEEGYPSTPG VRKALHVKRA PLQKRRVVQR STKSATAAC SLPTTAAPAQ LPLTSSGDAL TVTGGLSLQV TKQKRLVSYD VLDCTLGSGY HAGAVLENGG PYTRVVALDC D HDAMHAAR DLVEEFGGDR FRFYCCKMSE AKAMFGERSF DAIMIDGGVS DTQLEDPERG FLLDDEGGHR LDMRFGPQMG VG ALEYLNT VSQHTLVSSL LAYGLLEYGQ AMKMSRAITR RKPFVDSREV LTCIEQAGDE LPEGGWRSQG SRRKSPMSWK FLT SLRCII NNEMYELRQG IENALLMLRD DGRLVVFSRL PWEERLVRGT VDDHPHALLS YVEDISIDDV QIYGFTRHAK MWVI TRAAS SAYALKNTTT LTEEKFRESS VRWLTGMYAG QTHGFPANNF TFENFERKEW VTLRRNGKPP PVDVGLDDK UniProtKB: S-adenosyl-methyltransferase mraW-like protein, putative |
+Macromolecule #60: 9S rRNA
| Macromolecule | Name: 9S rRNA / type: rna / ID: 60 Details: 9S ribosomal RNA from Trypanosoma brucei. GenBank: X02547.1 and S66467.1 Sequence of 3'-major domain omitted. Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.505578 KDa |
| Sequence | String: UAAAUUAUGG UCAAUUGUUA GUAUUCAUAU UAAUUUUUUU AAAUGUUUUA UCAUUUUAUA AAGGUUUAUU UUUGAAAGAU UUUUUGUAU AAAAUUUUAG GAAUAGUUAA UAAUAAUUUA UAAUUUUGAU UAGAUUGUUU UGUUAAUGCU AUUAGAUGGG U GUGGAAAA ...String: UAAAUUAUGG UCAAUUGUUA GUAUUCAUAU UAAUUUUUUU AAAUGUUUUA UCAUUUUAUA AAGGUUUAUU UUUGAAAGAU UUUUUGUAU AAAAUUUUAG GAAUAGUUAA UAAUAAUUUA UAAUUUUGAU UAGAUUGUUU UGUUAAUGCU AUUAGAUGGG U GUGGAAAA AUAAAAAAAA UAAUUAAUAU AUAUCAAUAA UAAAUUAAAU UAAUCUAUUA GUCAGAAAUG GAUGCCAGCC GU UGCGGUA AUUUCUAUGC UUUUAAAUAU UAUACAAUUA UCAUAUUAAA UUGUUAAGUG CUGAUUUAAC CAAUAAAAAU AUA AAUAAU UUUUAUUUGU UUUUAAACAC CAUUAGGUAU AUGCAAAUAU AAAAUUAUAG UAAUUAUAGA AAUUAAAAAG GUAU UGUUG CCCACCAAUU UUUAUAAUAA AAAUAACGUG CAGUAAUUAA UAUAUUUAUA AAAAUAUAUU UUUUUUUU(UBD) |
+Macromolecule #63: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 63 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #64: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 64 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
+Macromolecule #65: S-(2-{[N-(2-HYDROXY-4-{[HYDROXY(OXIDO)PHOSPHINO]OXY}-3,3-DIMETHYL...
| Macromolecule | Name: S-(2-{[N-(2-HYDROXY-4-{[HYDROXY(OXIDO)PHOSPHINO]OXY}-3,3-DIMETHYLBUTANOYL)-BETA-ALANYL]AMINO}ETHYL) DECANETHIOATE type: ligand / ID: 65 / Number of copies: 1 / Formula: PM8 |
|---|---|
| Molecular weight | Theoretical: 496.598 Da |
| Chemical component information |  ChemComp-PM8: |
+Macromolecule #66: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 66 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
| Details | Body domain of the mitochondrial ribosomal small subunit assemblosome of Trypanosoma brucei brucei Lister strain 427 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 100719 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER Details: Startup model calculated from a small negative stain data set using EMD-0230 (T.brucei mitoribosomal small subunit) as an initial reference for classification of mitochondrial small subunit-like classes |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 161661 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
-Atomic model buiding 1
| Details | Starting models of ribosomal proteins of the T. brucei mitochondrial ribosomal small subunit were taken from PDB 6HIW. Mitchondrial ribosomal small subunit assembly factors were built de novo. |
|---|---|
| Refinement | Protocol: OTHER |
| Output model |  PDB-6sga: |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)