[English] 日本語
 Yorodumi
Yorodumi- EMDB-10131: MKLP2-decocrated 13 protofilament microtubule (ADP.Al.Fx state) p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10131 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | MKLP2-decocrated 13 protofilament microtubule (ADP.Al.Fx state) processed with MiRP (Microtubule Relion-based Pipeline) | ||||||||||||||||||
 Map data Map data | MKLP2 (ADPAlFx state) decorated 13PF microtubule: symmetrised asymmetric unit density from MiRP processing | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |   | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||||||||
 Authors Authors | Cook AC / Manka SW / Wang S / Moores CA / Atherton J | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. Authors: Joseph Atherton / I-Mei Yu / Alexander Cook / Joseph M Muretta / Agnel Joseph / Jennifer Major / Yannick Sourigues / Jeffrey Clause / Maya Topf / Steven S Rosenfeld / Anne Houdusse / Carolyn A Moores /    Abstract: MKLP2, a kinesin-6, has critical roles during the metaphase-anaphase transition and cytokinesis. Its motor domain contains conserved nucleotide binding motifs, but is divergent in sequence (~35% ...MKLP2, a kinesin-6, has critical roles during the metaphase-anaphase transition and cytokinesis. Its motor domain contains conserved nucleotide binding motifs, but is divergent in sequence (~35% identity) and size (~40% larger) compared to other kinesins. Using cryo-electron microscopy and biophysical assays, we have undertaken a mechanochemical dissection of the microtubule-bound MKLP2 motor domain during its ATPase cycle, and show that many facets of its mechanism are distinct from other kinesins. While the MKLP2 neck-linker is directed towards the microtubule plus-end in an ATP-like state, it does not fully dock along the motor domain. Furthermore, the footprint of the MKLP2 motor domain on the MT surface is altered compared to motile kinesins, and enhanced by kinesin-6-specific sequences. The conformation of the highly extended loop6 insertion characteristic of kinesin-6s is nucleotide-independent and does not contact the MT surface. Our results emphasize the role of family-specific insertions in modulating kinesin motor function. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10131.map.gz emd_10131.map.gz | 794.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10131-v30.xml emd-10131-v30.xml emd-10131.xml emd-10131.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10131.png emd_10131.png | 156 KB | ||
| Others |  emd_10131_additional.map.gz emd_10131_additional.map.gz emd_10131_additional_1.map.gz emd_10131_additional_1.map.gz | 285.2 MB 285.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10131 http://ftp.pdbj.org/pub/emdb/structures/EMD-10131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10131 | HTTPS FTP |
-Related structure data
| Related structure data |  4862C  6rf8C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10796 (Title: A microtubule RELION-based pipeline for cryo-EM image processing EMPIAR-10796 (Title: A microtubule RELION-based pipeline for cryo-EM image processingData size: 256.7 Data #1: Movies of microtubules decorated with MKLP2 motor domain (ADPAlFx bound) [micrographs - single frame] Data #2: Single-frames of microtubules decorated with MKLP2 motor domain (ADPAlFx bound) [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10131.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10131.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MKLP2 (ADPAlFx state) decorated 13PF microtubule: symmetrised asymmetric unit density from MiRP processing | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.534 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: MKLP2 (ADPAlFx state) decorated 13PF microtubule: C1 whole...
| File | emd_10131_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MKLP2 (ADPAlFx state) decorated 13PF microtubule: C1 whole microtubule density from MiRP processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
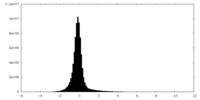
| Density Histograms |
-Additional map: MKLP2 (ADPAlFx state) decorated 13PF microtubule: C1 whole...
| File | emd_10131_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MKLP2 (ADPAlFx state) decorated 13PF microtubule: C1 whole microtubule density from MiRP processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
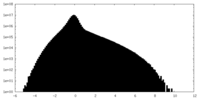
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of 13pf microtubule with bound MKLP2 motor domain in the ...
| Entire | Name: Complex of 13pf microtubule with bound MKLP2 motor domain in the presence of ADP.AlFx |
|---|---|
| Components |
|
-Supramolecule #1: Complex of 13pf microtubule with bound MKLP2 motor domain in the ...
| Supramolecule | Name: Complex of 13pf microtubule with bound MKLP2 motor domain in the presence of ADP.AlFx type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: Microtubule bound to taxol |
|---|
-Supramolecule #2: Tubulin alpha and beta chains
| Supramolecule | Name: Tubulin alpha and beta chains / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 / Details: Microtubule bound to taxol |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Kinesin-like protein KIF20A
| Supramolecule | Name: Kinesin-like protein KIF20A / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 / Details: Microtubule bound to taxol |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)