+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10041 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SIVrcm intasome | |||||||||
 Map data Map data | map sharpened with B factor -170 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | retroviral integrase / lentivirus / strand transfer inhibior / protein-DNA complex / RECOMBINATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationexoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / host cell / viral nucleocapsid ...exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / host cell / viral nucleocapsid / DNA recombination / aspartic-type endopeptidase activity / host cell cytoplasm / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / host cell nucleus / host cell plasma membrane / proteolysis / DNA binding / zinc ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Simian immunodeficiency virus Simian immunodeficiency virus | |||||||||
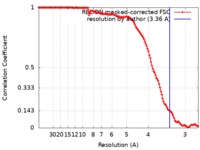
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Cherepanov P / Nans A / Cook N | |||||||||
| Funding support |  United States, United States,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis of second-generation HIV integrase inhibitor action and viral resistance. Authors: Nicola J Cook / Wen Li / Dénes Berta / Magd Badaoui / Allison Ballandras-Colas / Andrea Nans / Abhay Kotecha / Edina Rosta / Alan N Engelman / Peter Cherepanov /    Abstract: Although second-generation HIV integrase strand-transfer inhibitors (INSTIs) are prescribed throughout the world, the mechanistic basis for the superiority of these drugs is poorly understood. We ...Although second-generation HIV integrase strand-transfer inhibitors (INSTIs) are prescribed throughout the world, the mechanistic basis for the superiority of these drugs is poorly understood. We used single-particle cryo-electron microscopy to visualize the mode of action of the advanced INSTIs dolutegravir and bictegravir at near-atomic resolution. Glutamine-148→histidine (Q148H) and glycine-140→serine (G140S) amino acid substitutions in integrase that result in clinical INSTI failure perturb optimal magnesium ion coordination in the enzyme active site. The expanded chemical scaffolds of second-generation compounds mediate interactions with the protein backbone that are critical for antagonizing viruses containing the Q148H and G140S mutations. Our results reveal that binding to magnesium ions underpins a fundamental weakness of the INSTI pharmacophore that is exploited by the virus to engender resistance and provide a structural framework for the development of this class of anti-HIV/AIDS therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10041.map.gz emd_10041.map.gz | 164.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10041-v30.xml emd-10041-v30.xml emd-10041.xml emd-10041.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10041_fsc.xml emd_10041_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10041.png emd_10041.png | 66.8 KB | ||
| Masks |  emd_10041_msk_1.map emd_10041_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10041.cif.gz emd-10041.cif.gz | 7 KB | ||
| Others |  emd_10041_additional_1.map.gz emd_10041_additional_1.map.gz emd_10041_additional_2.map.gz emd_10041_additional_2.map.gz emd_10041_half_map_1.map.gz emd_10041_half_map_1.map.gz emd_10041_half_map_2.map.gz emd_10041_half_map_2.map.gz | 89.4 MB 167.9 MB 164.7 MB 164.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10041 http://ftp.pdbj.org/pub/emdb/structures/EMD-10041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10041 | HTTPS FTP |
-Related structure data
| Related structure data |  6rwlMC  6rwmC  6rwnC  6rwoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10041.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10041.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map sharpened with B factor -170 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10041_msk_1.map emd_10041_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
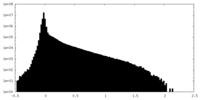
| Density Histograms |
-Additional map: Unsharpened CryoSPARC map
| File | emd_10041_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened CryoSPARC map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Autosharpened map from CryoSPARC
| File | emd_10041_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Autosharpened map from CryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
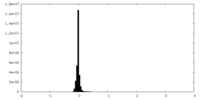
| Density Histograms |
-Half map: Half map A
| File | emd_10041_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
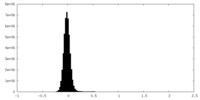
| Density Histograms |
-Half map: Half map B
| File | emd_10041_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
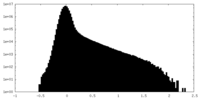
| Density Histograms |
- Sample components
Sample components
-Entire : SIVrcm intasome in complex with bictegravir
| Entire | Name: SIVrcm intasome in complex with bictegravir |
|---|---|
| Components |
|
-Supramolecule #1: SIVrcm intasome in complex with bictegravir
| Supramolecule | Name: SIVrcm intasome in complex with bictegravir / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: Pol Protein
| Supramolecule | Name: Pol Protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
-Macromolecule #1: Pol protein
| Macromolecule | Name: Pol protein / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 32.732182 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GFLDGIEKAQ EEHEKYHNNW RAMAEDFQIP QVVAKEIVAQ CPKCQVKGEA MHGQVDASPK TWQMDCTHLE GKVIIVAVHV ASGYIEAEV LPAETGKETA HFLLKLAARW PVKHLHTDNG DNFTSSAVQA VCWWAQIEHT FGVPYNPQSQ GVVESMNHQL K TIITQIRD ...String: GFLDGIEKAQ EEHEKYHNNW RAMAEDFQIP QVVAKEIVAQ CPKCQVKGEA MHGQVDASPK TWQMDCTHLE GKVIIVAVHV ASGYIEAEV LPAETGKETA HFLLKLAARW PVKHLHTDNG DNFTSSAVQA VCWWAQIEHT FGVPYNPQSQ GVVESMNHQL K TIITQIRD QAEKIETAVQ MAVLIHNFKR KGGIGGYSAG ERIIDIIASD LQTTKLQNQI SKIQNFRVYF REGRDQQWKG PA TLIWKGE GAVVIQDGQD LKVVPRRKCK IIKDYGRKDV DSETSMEGRQ EKD UniProtKB: Gag-Pol polyprotein |
-Macromolecule #2: Pol protein
| Macromolecule | Name: Pol protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 32.675131 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FLDGIEKAQE EHEKYHNNWR AMAEDFQIPQ VVAKEIVAQC PKCQVKGEAM HGQVDASPKT WQMDCTHLEG KVIIVAVHVA SGYIEAEVL PAETGKETAH FLLKLAARWP VKHLHTDNGD NFTSSAVQAV CWWAQIEHTF GVPYNPQSQG VVESMNHQLK T IITQIRDQ ...String: FLDGIEKAQE EHEKYHNNWR AMAEDFQIPQ VVAKEIVAQC PKCQVKGEAM HGQVDASPKT WQMDCTHLEG KVIIVAVHVA SGYIEAEVL PAETGKETAH FLLKLAARWP VKHLHTDNGD NFTSSAVQAV CWWAQIEHTF GVPYNPQSQG VVESMNHQLK T IITQIRDQ AEKIETAVQM AVLIHNFKRK GGIGGYSAGE RIIDIIASDL QTTKLQNQIS KIQNFRVYFR EGRDQQWKGP AT LIWKGEG AVVIQDGQDL KVVPRRKCKI IKDYGRKDVD SETSMEGRQE KD UniProtKB: Gag-Pol polyprotein |
-Macromolecule #3: Pol protein
| Macromolecule | Name: Pol protein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 32.68817 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GFLDGIEKAQ EEHEKYHNNW RAMAEDFQIP QVVAKEIVAQ CPKCQVKGEA MHGQVDASPK TWQMDCTHLE GKVIIVAVHV ASGYIEAEV LPAETGKETA HFLLKLAARW PVKHLHTDNG ANFTSSAVQA VCWWAQIEHT FGVPYNPQSQ GVVESMNHQL K TIITQIRD ...String: GFLDGIEKAQ EEHEKYHNNW RAMAEDFQIP QVVAKEIVAQ CPKCQVKGEA MHGQVDASPK TWQMDCTHLE GKVIIVAVHV ASGYIEAEV LPAETGKETA HFLLKLAARW PVKHLHTDNG ANFTSSAVQA VCWWAQIEHT FGVPYNPQSQ GVVESMNHQL K TIITQIRD QAEKIETAVQ MAVLIHNFKR KGGIGGYSAG ERIIDIIASD LQTTKLQNQI SKIQNFRVYF REGRDQQWKG PA TLIWKGE GAVVIQDGQD LKVVPRRKCK IIKDYGRKDV DSETSMEGRQ EKD UniProtKB: Gag-Pol polyprotein |
-Macromolecule #4: DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP...
| Macromolecule | Name: DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3') type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 9.223995 KDa |
| Sequence | String: (DG)(DT)(DT)(DC)(DT)(DA)(DG)(DA)(DA)(DG) (DG)(DC)(DT)(DA)(DA)(DG)(DA)(DA)(DA)(DA) (DA)(DT)(DC)(DT)(DC)(DT)(DA)(DC)(DC) (DA) |
-Macromolecule #5: DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*...
| Macromolecule | Name: DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3') type: dna / ID: 5 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 10.134541 KDa |
| Sequence | String: (DA)(DA)(DC)(DT)(DG)(DG)(DT)(DA)(DG)(DA) (DG)(DA)(DT)(DT)(DT)(DT)(DT)(DC)(DT)(DT) (DA)(DG)(DC)(DC)(DT)(DT)(DC)(DT)(DA) (DG)(DA)(DA)(DC) |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 8 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-30 / Number grids imaged: 2 / Number real images: 8027 / Average electron dose: 50.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)