+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0814 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
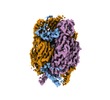
| Title | African swine fever virus major capsid protein p72 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | African Swine Fever virus capsomer / Trisymmetron / NCLDV / VIRAL PROTEIN | |||||||||
| Function / homology | Major capsid protein, C-terminal / Major capsid protein, C-terminal domain superfamily / Large eukaryotic DNA virus major capsid protein / Group II dsDNA virus coat/capsid protein / viral capsid / structural molecule activity / B646L Function and homology information Function and homology information | |||||||||
| Biological species |   African swine fever virus African swine fever virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Wang N / Rao Z | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Architecture of African swine fever virus and implications for viral assembly. Authors: Nan Wang / Dongming Zhao / Jialing Wang / Yangling Zhang / Ming Wang / Yan Gao / Fang Li / Jingfei Wang / Zhigao Bu / Zihe Rao / Xiangxi Wang /  Abstract: African swine fever virus (ASFV) is a giant and complex DNA virus that causes a highly contagious and often lethal swine disease for which no vaccine is available. Using an optimized image ...African swine fever virus (ASFV) is a giant and complex DNA virus that causes a highly contagious and often lethal swine disease for which no vaccine is available. Using an optimized image reconstruction strategy, we solved the ASFV capsid structure up to 4.1 angstroms, which is built from 17,280 proteins, including one major (p72) and four minor (M1249L, p17, p49, and H240R) capsid proteins organized into pentasymmetrons and trisymmetrons. The atomic structure of the p72 protein informs putative conformational epitopes, distinguishing ASFV from other nucleocytoplasmic large DNA viruses. The minor capsid proteins form a complicated network below the outer capsid shell, stabilizing the capsid by holding adjacent capsomers together. Acting as core organizers, 100-nanometer-long M1249L proteins run along each edge of the trisymmetrons that bridge two neighboring pentasymmetrons and form extensive intermolecular networks with other capsid proteins, driving the formation of the capsid framework. These structural details unveil the basis of capsid stability and assembly, opening up new avenues for African swine fever vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0814.map.gz emd_0814.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0814-v30.xml emd-0814-v30.xml emd-0814.xml emd-0814.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0814.png emd_0814.png | 102.7 KB | ||
| Filedesc metadata |  emd-0814.cif.gz emd-0814.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0814 http://ftp.pdbj.org/pub/emdb/structures/EMD-0814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0814 | HTTPS FTP |
-Related structure data
| Related structure data |  6l2tMC  0810C  0811C  0812C  0813C  0815C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0814.map.gz / Format: CCP4 / Size: 18.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0814.map.gz / Format: CCP4 / Size: 18.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : African swine fever virus
| Entire | Name:   African swine fever virus African swine fever virus |
|---|---|
| Components |
|
-Supramolecule #1: African swine fever virus
| Supramolecule | Name: African swine fever virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10497 / Sci species name: African swine fever virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: B646L,Major capsid protein
| Macromolecule | Name: B646L,Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   African swine fever virus African swine fever virus |
| Molecular weight | Theoretical: 73.219562 KDa |
| Sequence | String: MASGGAFCLI ANDGKADKII LAQDLLNSRI SNIKNVNKSY GKPDPEPTLS QIEETHLVHF NAHFKPYVPV GFEYNKVRPH TGTPTLGNK LTFGIPQYGD FFHDMVGHHI LGACHSSWQD APIQGTSQMG AHGQLQTFPR NGYDWDNQTP LEGAVYTLVD P FGRPIVPG ...String: MASGGAFCLI ANDGKADKII LAQDLLNSRI SNIKNVNKSY GKPDPEPTLS QIEETHLVHF NAHFKPYVPV GFEYNKVRPH TGTPTLGNK LTFGIPQYGD FFHDMVGHHI LGACHSSWQD APIQGTSQMG AHGQLQTFPR NGYDWDNQTP LEGAVYTLVD P FGRPIVPG TKNAYRNLVY YCEYPGLRLY ENVRFEVNGN SLDEYSSDVT TLVRKFCIPG DKMTGYKHLV GQEVSVEGTS GP LLCNIHD LHKPHQSKPI LTDENDTQRT CSHTNPKFLS QHFPHFPENI QTAGKQDITP ITDATYLDIR RNVHYSCNGP QTP KYYQPP LALWIKLRFW FNDNVNLAIP SVSIPFGERF ITIKLASQKD LVNEFPGLFV RQSTVIAGRP SRRNIRFKPW FIPG VINEI SLTNNELYIN NLFVTPEIHN LYVKRVRFSL IRVHKTQVTH TNNNHHDEKL MSALKWPIEY MFIGLKPTWN ISDQN PHQH RDWHKFGHVV NAIMQPTHHA EISFQDRDTA LPDACSSISD ISPVTYPITL PIIKNISVTA HGINLIDKFP SQFCSS YIP FHYGGNAIKT PDDPGAMMIT FALYPREEYQ PSGHINVSRA REFYISWDSD YVGSITTADL VVSSSAINFL LLQNGSA VL RYST |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















