[English] 日本語
 Yorodumi
Yorodumi- EMDB-0782: Cryo-EM structure of human IgM-Fc in complex with the J chain and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0782 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | |||||||||
 Map data Map data | The primary map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | immunoglobulin / pentamer / transcytosis / secreted / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / hexameric IgM immunoglobulin complex / polymeric immunoglobulin binding / dimeric IgA immunoglobulin complex / IgM B cell receptor complex / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / secretory IgA immunoglobulin complex ...polymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / hexameric IgM immunoglobulin complex / polymeric immunoglobulin binding / dimeric IgA immunoglobulin complex / IgM B cell receptor complex / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / secretory IgA immunoglobulin complex / Fc receptor signaling pathway / IgA binding / pre-B cell allelic exclusion / IgM immunoglobulin complex / glomerular filtration / detection of chemical stimulus involved in sensory perception of bitter taste / CD22 mediated BCR regulation / immunoglobulin receptor binding / azurophil granule membrane / receptor clustering / positive regulation of respiratory burst / humoral immune response / Scavenging of heme from plasma / antigen binding / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / epidermal growth factor receptor signaling pathway / transmembrane signaling receptor activity / antibacterial humoral response / protein-containing complex assembly / blood microparticle / protein-macromolecule adaptor activity / Potential therapeutics for SARS / defense response to Gram-negative bacterium / adaptive immune response / receptor complex / immune response / innate immune response / Neutrophil degranulation / cell surface / signal transduction / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
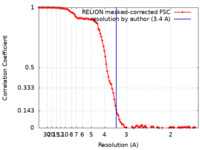
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Li Y / Wang G | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural insights into immunoglobulin M. Authors: Yaxin Li / Guopeng Wang / Ningning Li / Yuxin Wang / Qinyu Zhu / Huarui Chu / Wenjun Wu / Ying Tan / Feng Yu / Xiao-Dong Su / Ning Gao / Junyu Xiao /  Abstract: Immunoglobulin M (IgM) plays a pivotal role in both humoral and mucosal immunity. Its assembly and transport depend on the joining chain (J-chain) and the polymeric immunoglobulin receptor (pIgR), ...Immunoglobulin M (IgM) plays a pivotal role in both humoral and mucosal immunity. Its assembly and transport depend on the joining chain (J-chain) and the polymeric immunoglobulin receptor (pIgR), but the underlying molecular mechanisms of these processes are unclear. We report a cryo-electron microscopy structure of the Fc region of human IgM in complex with the J-chain and pIgR ectodomain. The IgM-Fc pentamer is formed asymmetrically, resembling a hexagon with a missing triangle. The tailpieces of IgM-Fc pack into an amyloid-like structure to stabilize the pentamer. The J-chain caps the tailpiece assembly and bridges the interaction between IgM-Fc and the polymeric immunoglobulin receptor, which undergoes a large conformational change to engage the IgM-J complex. These results provide a structural basis for the function of IgM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0782.map.gz emd_0782.map.gz | 14.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0782-v30.xml emd-0782-v30.xml emd-0782.xml emd-0782.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0782_fsc.xml emd_0782_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_0782.png emd_0782.png | 77.2 KB | ||
| Masks |  emd_0782_msk_1.map emd_0782_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0782.cif.gz emd-0782.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0782 http://ftp.pdbj.org/pub/emdb/structures/EMD-0782 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0782 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0782 | HTTPS FTP |
-Related structure data
| Related structure data |  6kxsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0782.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0782.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The primary map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0782_msk_1.map emd_0782_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of IgM-Fc with the J chain and pIgR/SC
| Entire | Name: Ternary complex of IgM-Fc with the J chain and pIgR/SC |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of IgM-Fc with the J chain and pIgR/SC
| Supramolecule | Name: Ternary complex of IgM-Fc with the J chain and pIgR/SC type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Immunoglobulin heavy constant mu
| Macromolecule | Name: Immunoglobulin heavy constant mu / type: protein_or_peptide / ID: 1 / Details: N-ACETYL-D-GLUCOSAMINE / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.875766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ASAWSHPQFE KGGGSGGGSG GSAWSHPQFE KIDTTIAELP PKVSVFVPPR DGFFGNPRKS KLICQATGFS PRQIQVSWLR EGKQVGSGV TTDQVQAEAK ESGPTTYKVT STLTIKESDW LGQSMFTCRV DHRGLTFQQN ASSMCVPDQD TAIRVFAIPP S FASIFLTK ...String: ASAWSHPQFE KGGGSGGGSG GSAWSHPQFE KIDTTIAELP PKVSVFVPPR DGFFGNPRKS KLICQATGFS PRQIQVSWLR EGKQVGSGV TTDQVQAEAK ESGPTTYKVT STLTIKESDW LGQSMFTCRV DHRGLTFQQN ASSMCVPDQD TAIRVFAIPP S FASIFLTK STKLTCLVTD LTTYDSVTIS WTRQNGEAVK THTNISESHP NATFSAVGEA SICEDDWNSG ERFTCTVTHT DL PSPLKQT ISRPKGVALH RPDVYLLPPA REQLNLRESA TITCLVTGFS PADVFVQWMQ RGQPLSPEKY VTSAPMPEPQ APG RYFAHS ILTVSEEEWN TGETYTCVVA HEALPNRVTE RTVDKSTGKP TLYNVSLVMS DTAGTCY UniProtKB: Immunoglobulin heavy constant mu |
-Macromolecule #2: Immunoglobulin J chain
| Macromolecule | Name: Immunoglobulin J chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.483329 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EDERIVLVDN KCKCARITSR IIRSSEDPNE DIVERNIRII VPLNNRENIS DPTSPLRTRF VYHLSDLCKK CDPTEVELDN QIVTATQSN ICDEDSATET CYTYDRNKCY TAVVPLVYGG ETKMVETALT PDACYPD UniProtKB: Immunoglobulin J chain |
-Macromolecule #3: Polymeric immunoglobulin receptor
| Macromolecule | Name: Polymeric immunoglobulin receptor / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.371875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KSPIFGPEEV NSVEGNSVSI TCYYPPTSVN RHTRKYWCRQ GARGGCITLI SSEGYVSSKY AGRANLTNFP ENGTFVVNIA QLSQDDSGR YKCGLGINSR GLSFDVSLEV SQGPGLLNDT KVYTVDLGRT VTINCPFKTE NAQKRKSLYK QIGLYPVLVI D SSGYVNPN ...String: KSPIFGPEEV NSVEGNSVSI TCYYPPTSVN RHTRKYWCRQ GARGGCITLI SSEGYVSSKY AGRANLTNFP ENGTFVVNIA QLSQDDSGR YKCGLGINSR GLSFDVSLEV SQGPGLLNDT KVYTVDLGRT VTINCPFKTE NAQKRKSLYK QIGLYPVLVI D SSGYVNPN YTGRIRLDIQ GTGQLLFSVV INQLRLSDAG QYLCQAGDDS NSNKKNADLQ VLKPEPELVY EDLRGSVTFH CA LGPEVAN VAKFLCRQSS GENCDVVVNT LGKRAPAFEG RILLNPQDKD GSFSVVITGL RKEDAGRYLC GAHSDGQLQE GSP IQAWQL FVNEESTIPR SPTVVKGVAG GSVAVLCPYN RKESKSIKYW CLWEGAQNGR CPLLVDSEGW VKAQYEGRLS LLEE PGNGT FTVILNQLTS RDAGFYWCLT NGDTLWRTTV EIKIIEGEPN LKVPGNVTAV LGETLKVPCH FPCKFSSYEK YWCKW NNTG CQALPSQDEG PSKAFVNCDE NSRLVSLTLN LVTRADEGWY WCGVKQGHFY GETAAVYVAV EERHHHHHHH H UniProtKB: Polymeric immunoglobulin receptor |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 59.74 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)