[English] 日本語
 Yorodumi
Yorodumi- EMDB-0666: Cdc48-Ufd1/Npl4 complex processing poly-ubiquitinated substrate i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0666 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cdc48-Ufd1/Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | ||||||||||||
 Map data Map data | Map sharpened with B factor of -150 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ATPase / ATPase complex / ubiquitin / quality control / MOTOR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationSCF complex disassembly in response to cadmium stress / mitotic DNA replication termination / Ovarian tumor domain proteases / Cdc48p-Npl4p-Vms1p AAA ATPase complex / Doa10p ubiquitin ligase complex / stress-induced homeostatically regulated protein degradation pathway / sister chromatid biorientation / protein localization to vacuole / endoplasmic reticulum membrane fusion / ribophagy ...SCF complex disassembly in response to cadmium stress / mitotic DNA replication termination / Ovarian tumor domain proteases / Cdc48p-Npl4p-Vms1p AAA ATPase complex / Doa10p ubiquitin ligase complex / stress-induced homeostatically regulated protein degradation pathway / sister chromatid biorientation / protein localization to vacuole / endoplasmic reticulum membrane fusion / ribophagy / Hrd1p ubiquitin ligase ERAD-L complex / DNA replication termination / RQC complex / mitochondria-associated ubiquitin-dependent protein catabolic process / positive regulation of mitochondrial fusion / cytoplasm protein quality control by the ubiquitin-proteasome system / HSF1 activation / protein-containing complex disassembly / nuclear protein quality control by the ubiquitin-proteasome system / protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / endosome to plasma membrane protein transport / protein phosphatase regulator activity / : / piecemeal microautophagy of the nucleus / mating projection tip / mitotic spindle disassembly / Protein methylation / VCP-NPL4-UFD1 AAA ATPase complex / replisome / ribosome-associated ubiquitin-dependent protein catabolic process / vesicle-fusing ATPase / K48-linked polyubiquitin modification-dependent protein binding / nuclear outer membrane-endoplasmic reticulum membrane network / retrograde protein transport, ER to cytosol / nonfunctional rRNA decay / KEAP1-NFE2L2 pathway / Neddylation / protein quality control for misfolded or incompletely synthesized proteins / polyubiquitin modification-dependent protein binding / autophagosome maturation / ribosomal large subunit export from nucleus / mRNA transport / ATP metabolic process / ERAD pathway / Neutrophil degranulation / rescue of stalled cytosolic ribosome / ubiquitin binding / macroautophagy / positive regulation of protein localization to nucleus / modification-dependent protein catabolic process / protein tag activity / ribosome biogenesis / ribosomal large subunit assembly / nuclear membrane / cytosolic large ribosomal subunit / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / cytoplasmic translation / structural constituent of ribosome / protein ubiquitination / ubiquitin protein ligase binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm / ATP hydrolysis activity / mitochondrion / ATP binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||
 Authors Authors | Twomey EC / Ji Z | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Authors: Edward C Twomey / Zhejian Ji / Thomas E Wales / Nicholas O Bodnar / Scott B Ficarro / Jarrod A Marto / John R Engen / Tom A Rapoport /  Abstract: The Cdc48 adenosine triphosphatase (ATPase) (p97 or valosin-containing protein in mammals) and its cofactor Ufd1/Npl4 extract polyubiquitinated proteins from membranes or macromolecular complexes for ...The Cdc48 adenosine triphosphatase (ATPase) (p97 or valosin-containing protein in mammals) and its cofactor Ufd1/Npl4 extract polyubiquitinated proteins from membranes or macromolecular complexes for subsequent degradation by the proteasome. How Cdc48 processes its diverse and often well-folded substrates is unclear. Here, we report cryo-electron microscopy structures of the Cdc48 ATPase in complex with Ufd1/Npl4 and polyubiquitinated substrate. The structures show that the Cdc48 complex initiates substrate processing by unfolding a ubiquitin molecule. The unfolded ubiquitin molecule binds to Npl4 and projects its N-terminal segment through both hexameric ATPase rings. Pore loops of the second ring form a staircase that acts as a conveyer belt to move the polypeptide through the central pore. Inducing the unfolding of ubiquitin allows the Cdc48 ATPase complex to process a broad range of substrates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0666.map.gz emd_0666.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0666-v30.xml emd-0666-v30.xml emd-0666.xml emd-0666.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0666.png emd_0666.png | 123.2 KB | ||
| Filedesc metadata |  emd-0666.cif.gz emd-0666.cif.gz | 6.6 KB | ||
| Others |  emd_0666_additional_1.map.gz emd_0666_additional_1.map.gz emd_0666_additional_2.map.gz emd_0666_additional_2.map.gz emd_0666_half_map_1.map.gz emd_0666_half_map_1.map.gz emd_0666_half_map_2.map.gz emd_0666_half_map_2.map.gz | 5.1 MB 24.4 MB 45.4 MB 45.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0666 http://ftp.pdbj.org/pub/emdb/structures/EMD-0666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0666 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0666 | HTTPS FTP |
-Related structure data
| Related structure data |  6oaaMC  0665C  6oa9C  6oabC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0666.map.gz / Format: CCP4 / Size: 48.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0666.map.gz / Format: CCP4 / Size: 48.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with B factor of -150 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
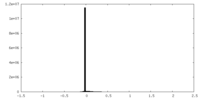
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Map sharpened with B factor of -100
| File | emd_0666_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with B factor of -100 | ||||||||||||
| Projections & Slices |
| ||||||||||||
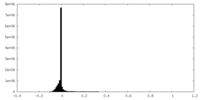
| Density Histograms |
-Additional map: Unsharpened map from refinement
| File | emd_0666_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
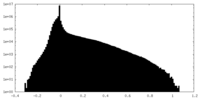
| Density Histograms |
-Half map: Half map 1 from refinement
| File | emd_0666_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
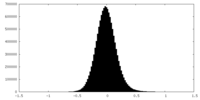
| Density Histograms |
-Half map: Half map 2 from refinement
| File | emd_0666_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the...
| Entire | Name: Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 |
|---|---|
| Components |
|
-Supramolecule #1: Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the...
| Supramolecule | Name: Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cell division control protein 48
| Macromolecule | Name: Cell division control protein 48 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: vesicle-fusing ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.106914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGEEHKPLLD ASGVDPREED KTATAILRRK KKDNMLLVDD AINDDNSVIA INSNTMDKLE LFRGDTVLVK GKKRKDTVLI VLIDDELED GACRINRVVR NNLRIRLGDL VTIHPCPDIK YATRISVLPI ADTIEGITGN LFDVFLKPYF VEAYRPVRKG D HFVVRGGM ...String: MGEEHKPLLD ASGVDPREED KTATAILRRK KKDNMLLVDD AINDDNSVIA INSNTMDKLE LFRGDTVLVK GKKRKDTVLI VLIDDELED GACRINRVVR NNLRIRLGDL VTIHPCPDIK YATRISVLPI ADTIEGITGN LFDVFLKPYF VEAYRPVRKG D HFVVRGGM RQVEFKVVDV EPEEYAVVAQ DTIIHWEGEP INREDEENNM NEVGYDDIGG CRKQMAQIRE MVELPLRHPQ LF KAIGIKP PRGVLMYGPP GTGKTLMARA VANETGAFFF LINGPEVMSK MAGESESNLR KAFEEAEKNA PAIIFIDEID SIA PKRDKT NGEVERRVVS QLLTLMDGMK ARSNVVVIAA TNRPNSIDPA LRRFGRFDRE VDIGIPDATG RLEVLRIHTK NMKL ADDVD LEALAAETHG YVGADIASLC SEAAMQQIRE KMDLIDLDED EIDAEVLDSL GVTMDNFRFA LGNSNPSALR ETVVE SVNV TWDDVGGLDE IKEELKETVE YPVLHPDQYT KFGLSPSKGV LFYGPPGTGK TLLAKAVATE VSANFISVKG PELLSM WYG ESESNIRDIF DKARAAAPTV VFLDELDSIA KARGGSLGDA GGASDRVVNQ LLTEMDGMNA KKNVFVIGAT NRPDQID PA ILRPGRLDQL IYVPLPDENA RLSILNAQLR KTPLEPGLEL TAIAKATQGF SGADLLYIVQ RAAKYAIKDS IEAHRQHE A EKEVKVEGED VEMTDEGAKA EQEPEVDPVP YITKEHFAEA MKTAKRSVSD AELRRYEAYS QQMKASRGQF SNFNFNDAP LGTTATDNAN SNNSAPSGAG AAFGSNAEED DDLYS UniProtKB: Cell division control protein 48 |
-Macromolecule #2: Nuclear protein localization protein 4
| Macromolecule | Name: Nuclear protein localization protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.862062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLIRFRSKNG THRVSCQEND LFGTVIEKLV GNLDPNADVD TFTVCEKPGQ GIHAVSELAD RTVMDLGLKH GDMLILNYSD KPANEKDGV NVEIGSVGID SKGIRQHRYG PLRIKELAVD EELEKEDGLI PRQKSKLCKH GDRGMCEYCS PLPPWDKEYH E KNKIKHIS ...String: MLIRFRSKNG THRVSCQEND LFGTVIEKLV GNLDPNADVD TFTVCEKPGQ GIHAVSELAD RTVMDLGLKH GDMLILNYSD KPANEKDGV NVEIGSVGID SKGIRQHRYG PLRIKELAVD EELEKEDGLI PRQKSKLCKH GDRGMCEYCS PLPPWDKEYH E KNKIKHIS FHSYLKKLNE NANKKENGSS YISPLSEPDF RINKRCHNGH EPWPRGICSK CQPSAITLQQ QEFRMVDHVE FQ KSEIINE FIQAWRYTGM QRFGYMYGSY SKYDNTPLGI KAVVEAIYEP PQHDEQDGLT MDVEQVKNEM LQIDRQAQEM GLS RIGLIF TDLSDAGAGD GSVFCKRHKD SFFLSSLEVI MAARHQTRHP NVSKYSEQGF FSSKFVTCVI SGNLEGEIDI SSYQ VSTEA EALVTADMIS GSTFPSMAYI NDTTDERYVP EIFYMKSNEY GITVKENAKP AFPVDYLLVT LTHGFPNTDT ETNSK FVSS TGFPWSNRQA MGQSQDYQEL KKYLFNVASS GDFNLLHEKI SNFHLLLYIN SLQILSPDEW KLLIESAVKN EWEESL LKL VSSAGWQTLV MILQESG UniProtKB: Nuclear protein localization protein 4 |
-Macromolecule #3: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.568769 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVESS DTIDNVKSKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRGG UniProtKB: Ubiquitin-ribosomal protein eL40A fusion protein |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 8 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: BERYLLIUM TRIFLUORIDE ION
| Macromolecule | Name: BERYLLIUM TRIFLUORIDE ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: BEF |
|---|---|
| Molecular weight | Theoretical: 66.007 Da |
| Chemical component information |  ChemComp-BEF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: GATAN CRYOPLUNGE 3 Details: Waited 20 seconds before blotting for 2.5-3 seconds.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 44.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 30118 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2) |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)