[English] 日本語
 Yorodumi
Yorodumi- EMDB-20730: Cryo-EM structure of p97-A232E mutant bound to cofactors UFD1L an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20730 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of p97-A232E mutant bound to cofactors UFD1L and NPLOC4 | |||||||||
 Map data Map data | Cryo-EM structure of the p97-A232E:NPLOC4-UFD1L complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
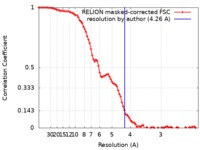
| Method | single particle reconstruction / cryo EM / Resolution: 4.26 Å | |||||||||
 Authors Authors | Blythe EE / Gates SN / Deshaies RJ / Martin A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: Multisystem Proteinopathy Mutations in VCP/p97 Increase NPLOC4·UFD1L Binding and Substrate Processing. Authors: Emily E Blythe / Stephanie N Gates / Raymond J Deshaies / Andreas Martin /  Abstract: Valosin-containing protein (VCP)/p97 is an essential ATP-dependent protein unfoldase. Dominant mutations in p97 cause multisystem proteinopathy (MSP), a disease affecting the brain, muscle, and bone. ...Valosin-containing protein (VCP)/p97 is an essential ATP-dependent protein unfoldase. Dominant mutations in p97 cause multisystem proteinopathy (MSP), a disease affecting the brain, muscle, and bone. Despite the identification of numerous pathways that are perturbed in MSP, the molecular-level defects of these p97 mutants are not completely understood. Here, we use biochemistry and cryoelectron microscopy to explore the effects of MSP mutations on the unfoldase activity of p97 in complex with its substrate adaptor NPLOC4⋅UFD1L (UN). We show that all seven analyzed MSP mutants unfold substrates faster. Mutant homo- and heterohexamers exhibit tighter UN binding and faster substrate processing. Our structural studies suggest that the increased UN affinity originates from a decoupling of p97's nucleotide state and the positioning of its N-terminal domains. Together, our data support a gain-of-function model for p97-UN-dependent processes in MSP and underscore the importance of N-terminal domain movements for adaptor recruitment and substrate processing by p97. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20730.map.gz emd_20730.map.gz | 7.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20730-v30.xml emd-20730-v30.xml emd-20730.xml emd-20730.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20730_fsc.xml emd_20730_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20730.png emd_20730.png | 211.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20730 http://ftp.pdbj.org/pub/emdb/structures/EMD-20730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20730 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20730.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20730.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the p97-A232E:NPLOC4-UFD1L complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of p97-A232E and cofactors UFD1L and NPLOC4
| Entire | Name: Complex of p97-A232E and cofactors UFD1L and NPLOC4 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of p97-A232E and cofactors UFD1L and NPLOC4
| Supramolecule | Name: Complex of p97-A232E and cofactors UFD1L and NPLOC4 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 191 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||||||||
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: Gatan Solarus Plasma Cleaner | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 2 second blot, 3 second wait. | |||||||||||||||||||||
| Details | Incubate complex for 5 minutes at 37 degrees Celsius before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 5897 / Average exposure time: 8.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)