+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-0654 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
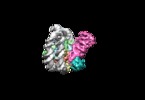
| タイトル | Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. 5.2A reconstruction of Dot1L on H2BK120Ub nucleosome | |||||||||
 マップデータ マップデータ | Result of cisTEM refinement, filtered to 5.2A, not sharpened. This map is in the same environment as the model constructed. It is shifted and resampled to fit with the environment of the paper | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 5.2 Å | |||||||||
 データ登録者 データ登録者 | Valencia-Sanchez MI / De Ioannes P / Wang M / Vasilyev N / Chen R / Nudler E / Armache J-P / Armache K-J | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2019 ジャーナル: Mol Cell / 年: 2019タイトル: Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. 著者: Marco Igor Valencia-Sánchez / Pablo De Ioannes / Miao Wang / Nikita Vasilyev / Ruoyu Chen / Evgeny Nudler / Jean-Paul Armache / Karim-Jean Armache /  要旨: The essential histone H3 lysine 79 methyltransferase Dot1L regulates transcription and genomic stability and is deregulated in leukemia. The activity of Dot1L is stimulated by mono-ubiquitination of ...The essential histone H3 lysine 79 methyltransferase Dot1L regulates transcription and genomic stability and is deregulated in leukemia. The activity of Dot1L is stimulated by mono-ubiquitination of histone H2B on lysine 120 (H2BK120Ub); however, the detailed mechanism is not understood. We report cryo-EM structures of human Dot1L bound to (1) H2BK120Ub and (2) unmodified nucleosome substrates at 3.5 Å and 4.9 Å, respectively. Comparison of both structures, complemented with biochemical experiments, provides critical insights into the mechanism of Dot1L stimulation by H2BK120Ub. Both structures show Dot1L binding to the same extended surface of the histone octamer. In yeast, this surface is used by silencing proteins involved in heterochromatin formation, explaining the mechanism of their competition with Dot1. These results provide a strong foundation for understanding conserved crosstalk between histone modifications found at actively transcribed genes and offer a general model of how ubiquitin might regulate the activity of chromatin enzymes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_0654.map.gz emd_0654.map.gz | 44.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-0654-v30.xml emd-0654-v30.xml emd-0654.xml emd-0654.xml | 17.1 KB 17.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_0654.png emd_0654.png | 70.6 KB | ||
| その他 |  emd_0654_additional.map.gz emd_0654_additional.map.gz emd_0654_half_map_1.map.gz emd_0654_half_map_1.map.gz emd_0654_half_map_2.map.gz emd_0654_half_map_2.map.gz | 59.3 MB 4.3 MB 4.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0654 http://ftp.pdbj.org/pub/emdb/structures/EMD-0654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0654 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0654 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_0654_validation.pdf.gz emd_0654_validation.pdf.gz | 79.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_0654_full_validation.pdf.gz emd_0654_full_validation.pdf.gz | 78.2 KB | 表示 | |
| XML形式データ |  emd_0654_validation.xml.gz emd_0654_validation.xml.gz | 493 B | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0654 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0654 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0654 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_0654.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_0654.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Result of cisTEM refinement, filtered to 5.2A, not sharpened. This map is in the same environment as the model constructed. It is shifted and resampled to fit with the environment of the paper | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
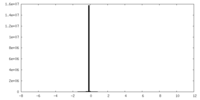
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-追加マップ: Result of cisTEM refinement, filtered to 5.2A, not...
| ファイル | emd_0654_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Result of cisTEM refinement, filtered to 5.2A, not sharpened. This map is not in the same environment as the model | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
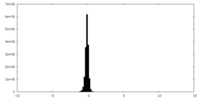
| 密度ヒストグラム |
-ハーフマップ: Half-map 1
| ファイル | emd_0654_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half-map 1 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
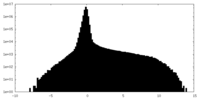
| 密度ヒストグラム |
-ハーフマップ: Half-map 2
| ファイル | emd_0654_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half-map 2 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
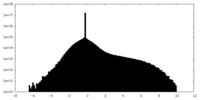
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Cryo-EM structure of human Dot1L bound to H2BK120Ub nucleosome at...
| 全体 | 名称: Cryo-EM structure of human Dot1L bound to H2BK120Ub nucleosome at 5.2A resolution |
|---|---|
| 要素 |
|
-超分子 #1: Cryo-EM structure of human Dot1L bound to H2BK120Ub nucleosome at...
| 超分子 | 名称: Cryo-EM structure of human Dot1L bound to H2BK120Ub nucleosome at 5.2A resolution タイプ: complex / ID: 1 / 親要素: 0 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 組換発現 | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| グリッド | 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 詳細: unspecified |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295.15 K / 装置: FEI VITROBOT MARK I 詳細: 3 ul of Dot1L-nucleosome complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 200 mesh), blotted in a Vitrobot Mark III (FEI Company) using 1.5 seconds ...詳細: 3 ul of Dot1L-nucleosome complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 200 mesh), blotted in a Vitrobot Mark III (FEI Company) using 1.5 seconds blotting at 100% humidity, and then plunge-frozen in liquid ethane cooled by liquid nitrogen.. |
| 詳細 | This sample was monodisperse |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: SUPER-RESOLUTION / デジタル化 - サンプリング間隔: 5.0 µm / 平均電子線量: 41.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT |
|---|
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)