[English] 日本語
 Yorodumi
Yorodumi- EMDB-0655: Structural basis of Dot1L stimulation by histone H2B lysine 120 u... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0655 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
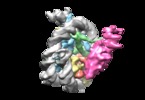
| Title | Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. 4.9A reconstruction of Dot1L on unmodified nucleosome | |||||||||
 Map data Map data | CryoSparc reconstruction, unsharpened. This density is rotated to fit with the final model. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Valencia-Sanchez MI / De Ioannes P / Wang M / Vasilyev N / Chen R / Nudler E / Armache J-P / Armache K-J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Authors: Marco Igor Valencia-Sánchez / Pablo De Ioannes / Miao Wang / Nikita Vasilyev / Ruoyu Chen / Evgeny Nudler / Jean-Paul Armache / Karim-Jean Armache /  Abstract: The essential histone H3 lysine 79 methyltransferase Dot1L regulates transcription and genomic stability and is deregulated in leukemia. The activity of Dot1L is stimulated by mono-ubiquitination of ...The essential histone H3 lysine 79 methyltransferase Dot1L regulates transcription and genomic stability and is deregulated in leukemia. The activity of Dot1L is stimulated by mono-ubiquitination of histone H2B on lysine 120 (H2BK120Ub); however, the detailed mechanism is not understood. We report cryo-EM structures of human Dot1L bound to (1) H2BK120Ub and (2) unmodified nucleosome substrates at 3.5 Å and 4.9 Å, respectively. Comparison of both structures, complemented with biochemical experiments, provides critical insights into the mechanism of Dot1L stimulation by H2BK120Ub. Both structures show Dot1L binding to the same extended surface of the histone octamer. In yeast, this surface is used by silencing proteins involved in heterochromatin formation, explaining the mechanism of their competition with Dot1. These results provide a strong foundation for understanding conserved crosstalk between histone modifications found at actively transcribed genes and offer a general model of how ubiquitin might regulate the activity of chromatin enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0655.map.gz emd_0655.map.gz | 30.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0655-v30.xml emd-0655-v30.xml emd-0655.xml emd-0655.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0655.png emd_0655.png | 123.2 KB | ||
| Others |  emd_0655_additional.map.gz emd_0655_additional.map.gz emd_0655_half_map_1.map.gz emd_0655_half_map_1.map.gz emd_0655_half_map_2.map.gz emd_0655_half_map_2.map.gz | 45.6 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0655 http://ftp.pdbj.org/pub/emdb/structures/EMD-0655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0655 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0655.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0655.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc reconstruction, unsharpened. This density is rotated to fit with the final model. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: CryoSparc reconstruction, mildly sharpened. This density is rotated...
| File | emd_0655_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc reconstruction, mildly sharpened. This density is rotated to fit with the final model. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoSparc half-map1. This map is in its original...
| File | emd_0655_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc half-map1. This map is in its original position, not in line with the final model | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoSparc half-map2. This map is in its original...
| File | emd_0655_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSparc half-map2. This map is in its original position, not in line with the final model | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of human Dot1L bound to unmodified nucleosome a...
| Entire | Name: Cryo-EM structure of human Dot1L bound to unmodified nucleosome at 4.9A resolution |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of human Dot1L bound to unmodified nucleosome a...
| Supramolecule | Name: Cryo-EM structure of human Dot1L bound to unmodified nucleosome at 4.9A resolution type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK I Details: 3 ul of Dot1L-nucleosome complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 200 mesh), blotted in a Vitrobot Mark III (FEI Company) using 1.5 seconds ...Details: 3 ul of Dot1L-nucleosome complexes were applied to a glow discharged Quantifoil holey carbon grid (1.2 um hole size, 200 mesh), blotted in a Vitrobot Mark III (FEI Company) using 1.5 seconds blotting at 100% humidity, and then plunge-frozen in liquid ethane cooled by liquid nitrogen.. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Sampling interval: 5.0 µm / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)