[English] 日本語
 Yorodumi
Yorodumi- EMDB-0570: HIV-1 vaccine elicited polyclonal 99-13 Fab in complex with the H... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0570 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
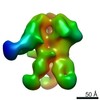
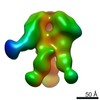
| Title | HIV-1 vaccine elicited polyclonal 99-13 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.5 Å | |||||||||
 Authors Authors | Nogal B / Ward AB | |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Authors: Kimberly M Cirelli / Diane G Carnathan / Bartek Nogal / Jacob T Martin / Oscar L Rodriguez / Amit A Upadhyay / Chiamaka A Enemuo / Etse H Gebru / Yury Choe / Federico Viviano / Catherine ...Authors: Kimberly M Cirelli / Diane G Carnathan / Bartek Nogal / Jacob T Martin / Oscar L Rodriguez / Amit A Upadhyay / Chiamaka A Enemuo / Etse H Gebru / Yury Choe / Federico Viviano / Catherine Nakao / Matthias G Pauthner / Samantha Reiss / Christopher A Cottrell / Melissa L Smith / Raiza Bastidas / William Gibson / Amber N Wolabaugh / Mariane B Melo / Benjamin Cossette / Venkatesh Kumar / Nirav B Patel / Talar Tokatlian / Sergey Menis / Daniel W Kulp / Dennis R Burton / Ben Murrell / William R Schief / Steven E Bosinger / Andrew B Ward / Corey T Watson / Guido Silvestri / Darrell J Irvine / Shane Crotty /   Abstract: Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the ...Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the immunological hurdles posed, including B cell immunodominance and germinal center (GC) quantity and quality. We found that two independent methods of slow delivery immunization of rhesus monkeys (RMs) resulted in more robust T follicular helper (T) cell responses and GC B cells with improved Env-binding, tracked by longitudinal fine needle aspirates. Improved GCs correlated with the development of >20-fold higher titers of autologous nAbs. Using a new RM genomic immunoglobulin locus reference, we identified differential IgV gene use between immunization modalities. Ab mapping demonstrated targeting of immunodominant non-neutralizing epitopes by conventional bolus-immunized animals, whereas slow delivery-immunized animals targeted a more diverse set of epitopes. Thus, alternative immunization strategies can enhance nAb development by altering GCs and modulating the immunodominance of non-neutralizing epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0570.map.gz emd_0570.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0570-v30.xml emd-0570-v30.xml emd-0570.xml emd-0570.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0570.png emd_0570.png | 48.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0570 http://ftp.pdbj.org/pub/emdb/structures/EMD-0570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0570 | HTTPS FTP |
-Validation report
| Summary document |  emd_0570_validation.pdf.gz emd_0570_validation.pdf.gz | 77.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0570_full_validation.pdf.gz emd_0570_full_validation.pdf.gz | 76.8 KB | Display | |
| Data in XML |  emd_0570_validation.xml.gz emd_0570_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0570 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0570 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0570 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0570 | HTTPS FTP |
-Related structure data
| Related structure data |  0569C  0571C  0572C  0573C  0574C  0575C  0576C  0577C  0578C  0579C  0580C  0581C  0582C  9138C  9175C  9176C  9177C  9178C  9179C  9180C  9181C  9182C  9183C  9184C  9185C  9186C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0570.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0570.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 vaccine elicited Fab in complex with the HIV Env trimer BG5...
| Entire | Name: HIV-1 vaccine elicited Fab in complex with the HIV Env trimer BG505 SOSIPv5.2 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 vaccine elicited Fab in complex with the HIV Env trimer BG5...
| Supramolecule | Name: HIV-1 vaccine elicited Fab in complex with the HIV Env trimer BG505 SOSIPv5.2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: SOSIP trimer
| Supramolecule | Name: SOSIP trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Polyclonal Fab
| Supramolecule | Name: Polyclonal Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















