+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0378 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | p53 dimer assembly | |||||||||
 Map data Map data | p53 dimer assembly | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Human (human) Human (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Kelly DF / Dearnaley WJ / Varano AC | |||||||||
 Citation Citation |  Journal: Small / Year: 2019 Journal: Small / Year: 2019Title: Cryo-EM-On-a-Chip: Custom-Designed Substrates for the 3D Analysis of Macromolecules. Authors: Nick A Alden / A Cameron Varano / William J Dearnaley / Maria J Solares / William Y Luqiu / Yanping Liang / Zhi Sheng / Sarah M McDonald / John Damiano / Jennifer McConnell / Madeline J ...Authors: Nick A Alden / A Cameron Varano / William J Dearnaley / Maria J Solares / William Y Luqiu / Yanping Liang / Zhi Sheng / Sarah M McDonald / John Damiano / Jennifer McConnell / Madeline J Dukes / Deborah F Kelly /  Abstract: The fight against human disease requires a multidisciplinary scientific approach. Applying tools from seemingly unrelated areas, such as materials science and molecular biology, researchers can ...The fight against human disease requires a multidisciplinary scientific approach. Applying tools from seemingly unrelated areas, such as materials science and molecular biology, researchers can overcome long-standing challenges to improve knowledge of molecular pathologies. Here, custom-designed substrates composed of silicon nitride (SiN) are used to study the 3D attributes of tumor suppressor proteins that function in DNA repair events. New on-chip preparation strategies enable the isolation of native protein complexes from human cancer cells. Combined techniques of cryo-electron microscopy (EM) and molecular modeling reveal a new modified form of the p53 tumor suppressor present in aggressive glioblastoma multiforme cancer cells. Taken together, the findings provide a radical new design for cryo-EM substrates to evaluate the structures of disease-related macromolecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0378.map.gz emd_0378.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0378-v30.xml emd-0378-v30.xml emd-0378.xml emd-0378.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0378.png emd_0378.png | 103.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0378 http://ftp.pdbj.org/pub/emdb/structures/EMD-0378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0378 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0378.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0378.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | p53 dimer assembly | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
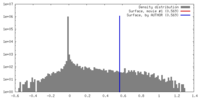
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : p53 dimer assembly
| Entire | Name: p53 dimer assembly |
|---|---|
| Components |
|
-Supramolecule #1: p53 dimer assembly
| Supramolecule | Name: p53 dimer assembly / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Brain / Tissue: Glioblastoma Homo sapiens (human) / Organ: Brain / Tissue: Glioblastoma |
| Molecular weight | Theoretical: 116 KDa |
-Macromolecule #1: p53
| Macromolecule | Name: p53 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human (human) / Organ: Brain / Tissue: Glioblastoma Human (human) / Organ: Brain / Tissue: Glioblastoma |
| Sequence | String: MEEPQSDPSV EPPLSQETFS DLWKLLPENN VLSPLPSQAM DDLMLSPDDI EQWFTEDPGP DEAPRMPEA APPVAPAPAA PTPAAPAPAP SWPLSSSVPS QKTYQGSYGF RLGFLHSGTA K SVTCTYSP ALNKMFCQLA KTCPVQLWVD STPPPGTRVR AMAIYKQSQH ...String: MEEPQSDPSV EPPLSQETFS DLWKLLPENN VLSPLPSQAM DDLMLSPDDI EQWFTEDPGP DEAPRMPEA APPVAPAPAA PTPAAPAPAP SWPLSSSVPS QKTYQGSYGF RLGFLHSGTA K SVTCTYSP ALNKMFCQLA KTCPVQLWVD STPPPGTRVR AMAIYKQSQH MTEVVRRCPH HE RCSDSDG LAPPQHLIRV EGNLRVEYLD DRNTFRHSVV VPYEPPEVGS DCTTIHYNYM CNS SCMGGM NRRPILTIIT LEDSSGNLLG RNSFEVRVCA CPGRDRRTEE ENLRKKGEPH HELP PGSTK RALPNNTSSS PQPKKKPLDG EYFTLQIRGR ERFEMFRELN EALELKDAQA GKEPG GSRA HSSHLKSKKG QSTSRHKKLM FKTEGPDSD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Component - Name: HEPES buffer Details: 20 mM HEPES, pH 7.2, 150 mM NaCl, 10 mM CaCl2, 10 mM MgCl2, 60 mM Imidazole |
| Grid | Material: SILICON NITRIDE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK III Details: Blot for 6-10 seconds before plunging, dwell time was 1 second, grid platform was SiN chip.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (2k x 2k) / Digitization - Sampling interval: 30.0 µm / Number grids imaged: 1 / Number real images: 100 / Average electron dose: 5.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus min: -1.5 µm / Nominal magnification: 68000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)